MaxWell Webinar with Dr. Hsiu-Chuan Lin
Tuesday, October 28, 2025 | 17:00 CET
09:00 PDT | 12:00 EDT | 00:00 CST | 01:00 JST
Webinar Hightlights
- Discover how transcription factor (TF) programming and morphogen signaling can be combined to generate diverse human neuronal subtypes in vitro.
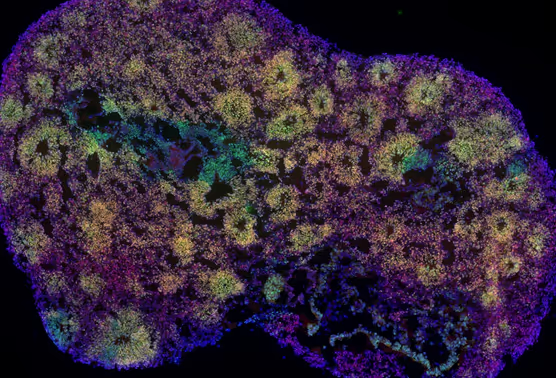
- Learn about a large-scale screen of 480 signaling conditions and over 700,000 single cells analyzed with multiplexed single-cell transcriptomics.
- Explore how MaxWell Biosystems’ MaxTwo HD-MEA platform enables high-resolution electrophysiological profiling, revealing distinct single-neuron properties, network activities, and morphological complexity.
- Understand how gene expression programs link to neuronal maturation and arborization, offering insights into neurodevelopment and disease modeling.
The webinar covered
- How combining TF overexpression with developmental signaling modulation enables the generation of diverse human neuron types.
- Findings from a comprehensive screen of 480 patterning conditions using single-cell transcriptomics on 700,000 cells.
- Functional profiling of neurons using MaxWell Biosystems’ MaxTwo HD-MEA platform to capture activity, connectivity, and morphological diversity across different patterning conditions.
- The relationship between transcriptomic signatures and neuronal structure, revealing mechanisms of maturation and identity formation.

Agenda
Profiling and programming in vitro neuronal diversity at single-cell resolution
Abstract
Human neurons programmed through transcription factor (TF) overexpression have revolutionized our ability to study human neurodevelopment and model neurological diseases, offering exciting opportunities for therapeutic screening and intervention. However, the diversity of neuronal subtypes programmable in vitro remains underexplored. Here, we modulate developmental signaling pathways combined with TF overexpression to explore the spectrum of neuron subtypes generated from pluripotent stem cells. We screened 480 morphogen signaling modulations coupled with NGN2 or ASCL1/DLX2 induction using a multiplexed single-cell transcriptomic readout. Analysis of 700,000 cells identified diverse excitatory and inhibitory neurons patterned along the developmental axes of the neural tube. Using electrophysiological measurements as functional readouts, we obtained highly reproducible and quantitative profiles that captured distinct single-neuron properties, network-level activities, and morphological features across different patterning conditions. Interestingly, the complexity of neuronal morphology was reflected in the transcriptome. Neurons with higher maturation scores exhibited increased arborization, suggesting a connection between gene expression programs and morphological maturation. Together, our work establishes a scalable framework for programming diverse human neuronal subtypes and offers a systematic approach to dissect how cooperative signaling drives neuronal identity and function.






.avif)

.avif)
.avif)


