
Disease Modeling
Every Cell has a Story to Tell.
Let’s Discover Yours
Disease modeling with in vitro cellular systems allows researchers to recreate and study the mechanisms of neurological disorders in controlled experimental settings.
In neuroscience and stem cell biology, this often involves patient-derived or genetically engineered neuronal models that capture key aspects of disease at the molecular, cellular, and network levels. These models can reveal pathological changes in excitability, synaptic transmission, and network dynamics, which are often linked to genetic mutations or environmental insults. By reproducing disease-relevant functional and structural abnormalities, in vitro systems provide a powerful platform for dissecting disease mechanisms, identifying biomarkers, and evaluating candidate therapeutics. Combined with electrophysiology, imaging, and multi-omics approaches, these models offer deeper insights into how dysfunction emerges and progresses in the nervous system.
MaxWell Biosystems’ HD-MEA platforms bring powerful capabilities to in vitro disease modeling by enabling detailed, label-free recordings of pathological activity across single neurons and entire networks. With unmatched signal fidelity and resolution, researchers can detect subtle dysfunctions, quantify altered phenotypes, and monitor disease progression or therapeutic rescue effects over time. This enables researchers to link cellular dysfunction to disease mechanisms and establish in vitro assays for testing therapeutic strategies in a human-relevant context.
Have a look at our Focus page about Neurological Disease Modeling.

Reliable disease signatures through high reproducibility
Disease models often display variable phenotypes across cell lines or mutations. Reproducible network activity across wells and timepoints helps distinguish true pathological features from noise or sampling artifacts, providing reliable insight into disease-related dysfunction.
Reveal disease-driven changes in axonal function
Many neurological disorders involve impaired signal transmission and disrupted connectivity. High-resolution axon tracking enables measurement of conduction velocity and axonal morphology, helping researchers identify functional connectivity deficits and altered axonal growth linked to disease pathology.
Detect subtle signs of dysfunction at earlier stages
Disease phenotypes may appear as subtle changes in spiking, network behavior, or excitability. Sensitive detection of low-amplitude and sparse signals enables identification of early-stage dysfunction, even before noticeable degeneration or structural changes emerge.
Scale up your disease model studies
Testing multiple patient-derived lines or disease conditions requires high-throughput, consistent analysis. Our automation-ready HD-MEA platforms ensure robust data acquisition and processing across wells and timepoints, enabling efficient screening of phenotypes and therapeutic responses.

Case studies
Sensitive detection of disease-related network phenotype in dopaminergic neuronal models
This study utilized the MaxTwo multi-well HD-MEA system to characterize iPSC-derived dopaminergic neurons with 16p11.2 duplication and deletion, genetic variants associated with schizophrenia, autism spectrum disorder and ADHD. Longitudinal recordings revealed pronounced hyperactivity and increased network synchrony in 16p11.2 deletion cultures compared to controls and duplication lines. MaxWell Biosystems’ platform provided sensitive and highly reproducible detection of genotype-specific network phenotypes, enabling robust identification of disease-related functional changes over time. Read more in Sundberg et al., Nat. Comm., 2021.
Disease-specific network activity in human iPSC-derived dopaminergic neurons
(a) Experimental timeline and (b,c) longitudinal analysis of active and synchronized sensor frequency (Hz) for control, 16p11.2 duplication, and 16p11.2 deletion neuron networks over 28 days. (d) Representative Activity Scan Assay activity maps visualizing overall neuronal activity at day 28 for each condition. (e) Synchronized sensor maps highlighting patterns of network synchrony in control, duplication, and deletion cultures.
Data adapted from Sundberg et al., Nature Communications, 2021.
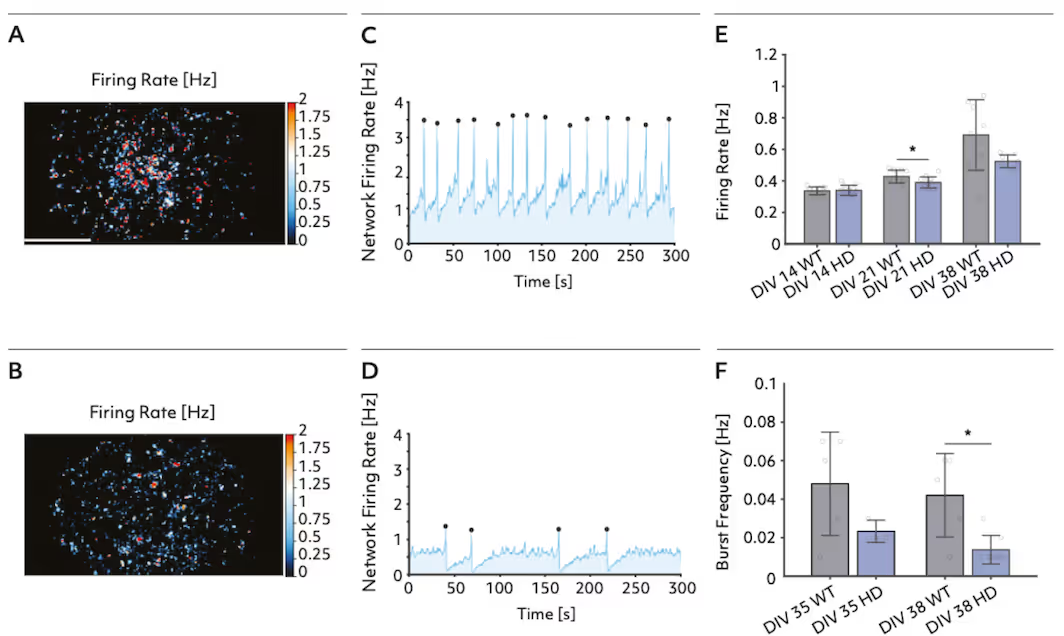
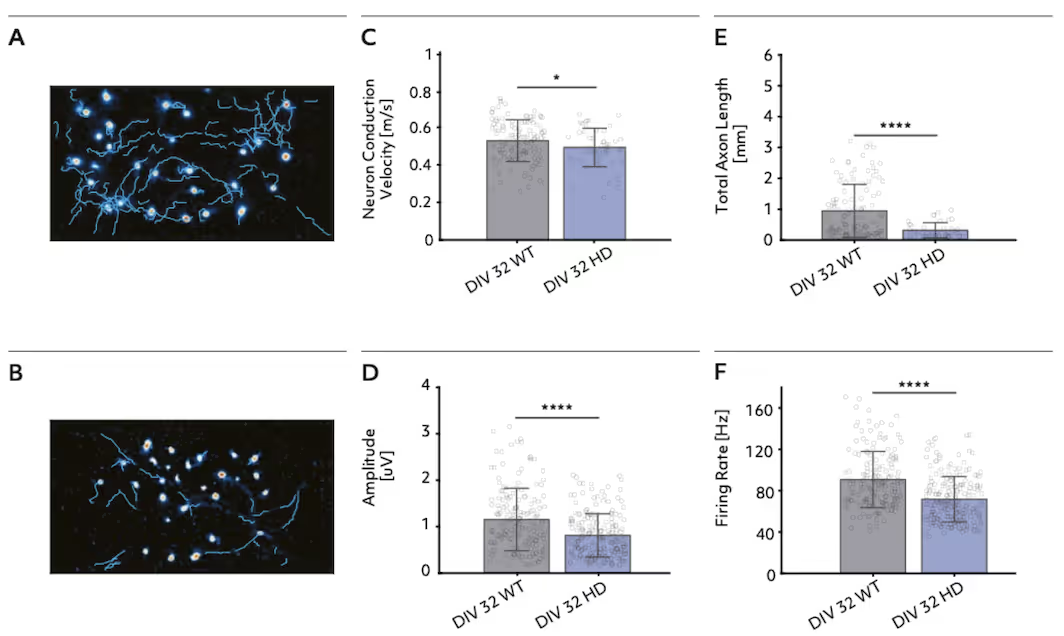
Axon morphology and network activity reveals Huntington's Disease phenotypes
This study leveraged the high spatial resolution of the MaxTwo multi-well HD-MEA platform to profile wild-type and Huntington’s disease (HD) model bit.bio ioGlutametergic Neurons at single-cell and network levels. The platforms’ sensitive Axon Tracking Assay enabled detection of distinct disease-related reductions in conduction velocity and total axon length. Comprehensive network analysis further revealed hypoactivity and reduced burst frequency in HD cultures, demonstrating robust and reproducible identification of functional phenotypes linked to neurodegenerative disease.
Axonal and network signatures in bit.bio ioGlutamergic HTT 50CAG/WT Huntington’s Disease neuron models
Top: Firing rate distribution maps and network activity traces show reduced neural activity and burst frequency in Huntington’s disease model neurons (HD) compared to wild type (WT) at DIV 38.
Bottom: Maps of action potential amplitude and quantitative analysis reveal marked declines in conduction velocity, total axon length, firing rate, and amplitude in HD neurons versus WT at DIV 32.
Data adapted from the Application Note in collaboration with bit.bio and Charles River Laboratories.
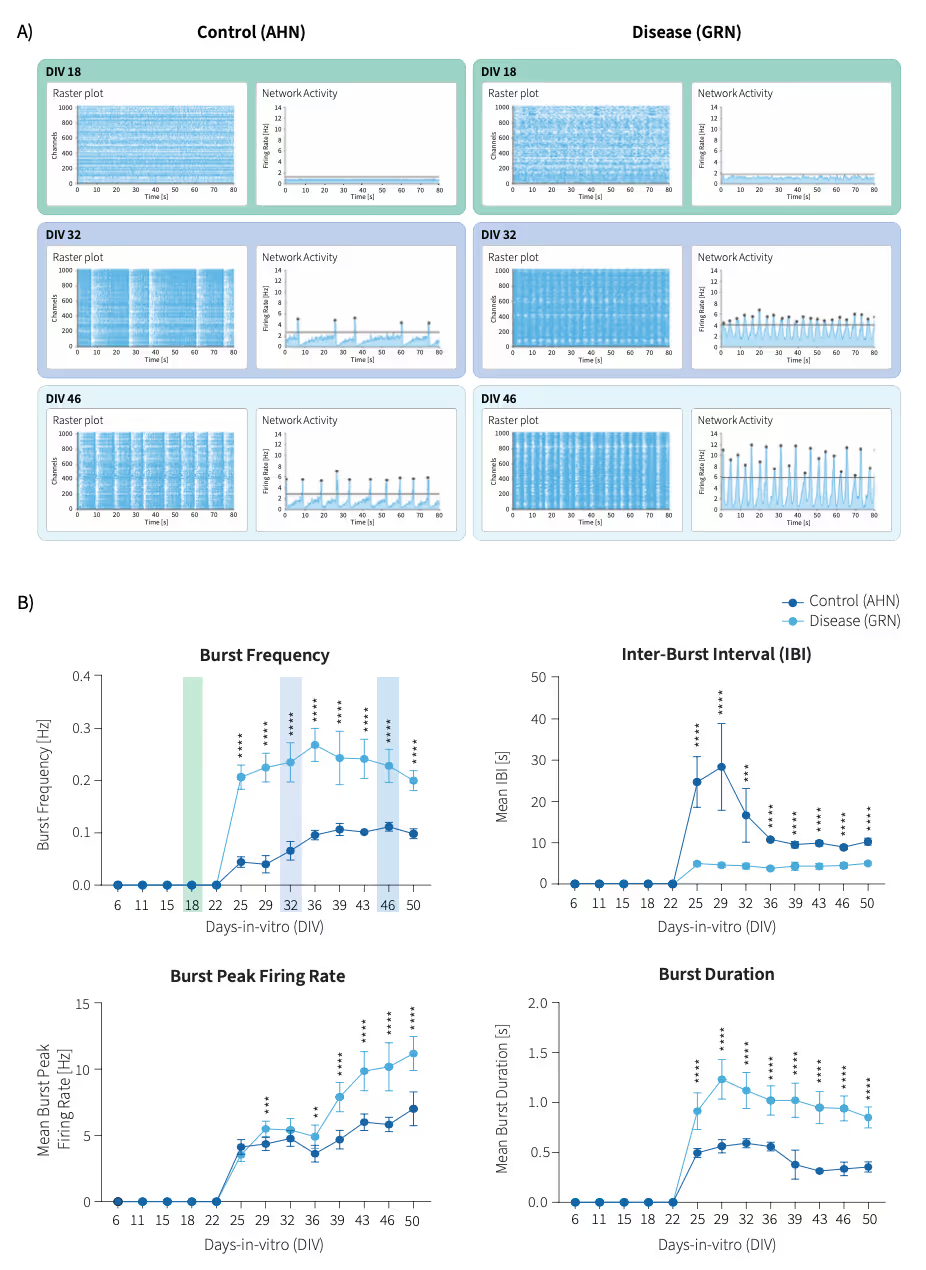
Reproducible detection of network hyperactivity in frontotemporal dementia models
This study utilized the MaxTwo 24-well HD-MEA system to characterize network dynamics in co-cultures of FUJIFILM Cellular Dynamics’ iCell Excitatory Induced Neurons and iCell Astrocytes 2.0 modeling frontotemporal dementia (FTD). Longitudinal recordings uncovered hallmark features of hyperactivity and disorganized network activity in the FTD disease line compared to controls. This platform enabled sensitive, reproducible quantification of multiple network parameters, providing robust identification of disease-relevant functional phenotypes for neurodegenerative research.
Consistent Network Phenotypes in Frontotemporal Dementia Neuron Models
Top: Representative raster and network activity plots at DIV 18, 32, and 46 illustrate reproducible differences in activity and burst structure between FTD (featuring a heterozygous knockout mutation in the progranulin (GRN) gene) and control (AHN) cultures.
Bottom: Longitudinal quantification of burst frequency, inter-burst interval, peak firing rate, and burst duration highlights persistently enhanced excitability and altered network dynamics in the FTD line across wells and time.
Data adapted from the Application note in collaboration with Fujifilm Cellular Dynamics, Inc.
Selected Resources
Axonal Eif5a hypusination controls local translation and mitigates defects in FUS-ALS

Human iPSC-derived glutamatergic neurons with pathogenic KCNQ2 variants display hyperactive bursting phenotypes

The fast-dissociating D2 antagonist antipsychotic JNJ-37822681 is a neuronal Kv7 channel opener: Potential repurposing for epilepsy treatment

A model of human neural networks reveals NPTX2 pathology in ALS and FTLD

Multiplex epigenome editing of MECP2 to rescue Rett syndrome neurons

Application Note with CRL and bit.bio
Developing next-generation in-vitro phenotypic assays for Huntington's disease by combining precision reprogrammed hiPSC-derived disease models with high-density microelectrode arrays.
MxW - FCDI Application Note
Longitudinal Functional Profiling of HumaniPSC-derived Frontotemporal Dementia Neuronson HD-MEAs




.avif)


