Microphysiological Systems
Every Cell has a Story to Tell.
Let’s Discover Yours
Microphysiological systems (MPS), such as controlled neuronal networks using microfluidics, microtunnels, and compartmentalization, provide advanced in vitro models for studying complex brain functions in a structured and reproducible manner.
These systems recreate key features of neural circuitry, enabling precise control over cell placement, connectivity, and signal flow. As a result, they are increasingly used to investigate neurodevelopment, circuit-level function, disease mechanisms, and compound responses in a human-relevant context.
MaxWell Biosystems’ High-Density Microelectrode Array (HD-MEA) platforms are uniquely suited for use with microphysiological systems. The HD-MEA’s ultra-high sensitivity captures every signal, from individual action potentials to axonal propagation, ensuring no functional activity is missed. This makes MaxWell Biosystems’ HD-MEAs a powerful platform for probing and understanding complex, structured neuronal dynamics in microphysiological systems.

Build networks on a flat, MPS-compatible surface
The flat planar surface of MaxWell Biosystems’ HD-MEAs supports easy integration of compartmentalized systems and chip-based platforms, whether PDMS-based or other materials. With this MPS-compatible surface, structures remain well attached to the HD-MEA surface for longitudinal experiments.
Faster setup – high electrode coverage eliminates the need for fine alignment
With dense, full-area electrode coverage, functional regions of MPS structures including axonal tunnels are reliably captured without the need for micrometer-level alignment. Researchers can simply place the MPS structures, culture the cells, and begin recording. This streamlined setup reduces preparation time and enables efficient scaling of MPS-based experiments.
Know exactly where to record and stimulate
Easily identify which electrodes are exposed to cells within the MPS structures, enabling targeted stimulation and recording from specific compartments.
Detect every cell, every axon, every signal
Capture spontaneous and evoked activity with subcellular resolution, including axonal propagation and low-amplitude signals across channels, bridges, and microtunnels. This high sensitivity supports consistent, reproducible results across experiments.
Stimulate with precision
Deliver localized electrical stimulation using individually addressable electrodes, ideal for probing connectivity, synaptic strength, and circuit responses within defined regions of the MPS.

Case studies
Engineered neural networks on HD-MEA facilitate node-specific functionality and connectivity
This study showcases a bottom‑up neuroscience platform that builds up to six separate neural circuits of primary rat neurons on flat, planar MaxOne+ HD-MEA Chips. Compact PDMS microstructures enable construction of topologically defined compartmentalized networks, suited for precise investigation of neuronal activity in reduced‑complexity systems. Primary rat neurons grown inside these microstructures on HD-MEAs yield networks whose activity can be recorded with subcellular resolution, allowing detailed mapping of functional connectivity and signal propagation. Read more in Duru et al., Biosensors and Bioelectronics, 2023.
Primary rat neurons grown inside PDMS microstructures on HD-MEAs yield topologically constrained neural networks
Primary rat cortical neurons are patterned within PDMS microstructures placed on HD-MEA chips, forming isolated neural networks suitable for precise electrophysiological analysis. (B) Confocal image at DIV 8 shows discrete cell clusters within microstructures. (Ci) Scanning electron micrograph (SEM) depicts a network node on the electrode grid at DIV 8; (Cii) false-colored SEM image highlights axonal bundles confined inside microchannels. Electrode coverage supports recording of activity at subcellular resolution within defined compartments.
Data adapted from Duru et al., Biosensors and Bioelectronics, 2023.
Microphysiological system for mapping sensory-spinal neuron interactions
This study introduces a sensory‑spinal co‑culture microphysiological model that reveals how dorsal‑root‑ganglion sensory neurons can drive sustained activation of spinal neurons, a hallmark of nociplastic pain. Targeted optical stimulation on the MaxOne HD-MEA Chip captured long-lasting, plasticity-linked spinal responses, modeling sensory-driven spinal dynamics in vitro. The platform’s high sensitivity enables detection of every signal, including subcellular events and low-amplitude activity across microtunnels. Read more in Miyahara et al., Frontiers in Neuroscience, 2025.
Microphysiological System of Sensory-Spinal Neuron Co-Culture on HD-MEAs
Left: Schematic shows PDMS microstructures positioned on HD-MEA chips, enabling separate culture of DRG and spinal neurons connected by microtunnels. Optical stimulation is used to activate individual DRG neurons expressing Channelrhodopsin-2, while spinal neuron responses are recorded.
Right: Immunofluorescence images illustrate peripherin+ DRG neurons (red), NeuN+ spinal neurons (green), and merged overlays visualizing axonal projections and GFP+ cells, supporting compartment-specific stimulation and analysis.
Data adapted from Miyahara et al., Frontiers in Neuroscience, 2025.
Mapping functional connectivity in modular and random neuronal networks
This study examines spontaneous activity and functional connectivity in engineered modular and random networks cultured on HD-MEA chips. PDMS microfluidic geometry defines modular structures, allowing direct comparison of compartmentalized and distributed network interactions through high-resolution correlation mapping. The dense, full-area electrode coverage enables reliable capture of functional regions, including microcompartments, without the need for fine alignment. Read more in Sato et al., Frontiers in Neuroscience, 2023.
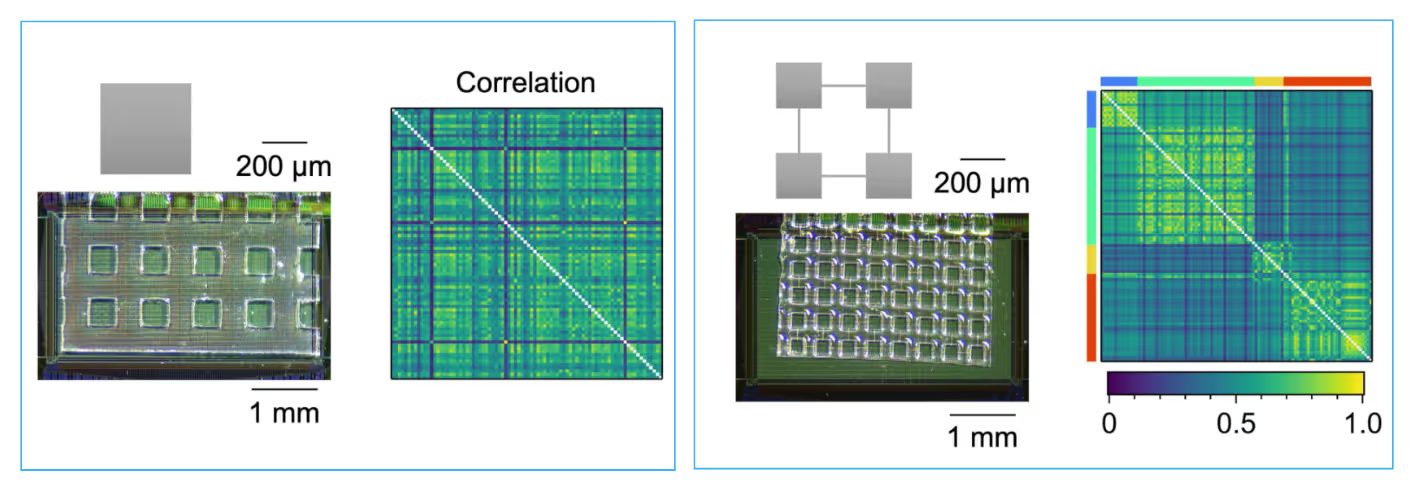
Spatial Patterns and Connectivity in Microstructured Networks
Spontaneous activity in random networks (Left) and modular networks (Right) at DIV 14. Microscopy images show the PDMS microfluidic film and network geometry on the HD-MEA. Correlation matrices illustrate network structures, where random networks (Left) exhibit broader distributions, while modular networks (Right) display compartmentalized connectivity patterns.
Data adapted from Sato et al., Frontiers in Neuroscience, 2023.
Custom MPS solutions for your HD-MEA research
If you are interested in developing a custom microphysiological system (MPS) structure on MaxWell Biosystems' HD-MEAs and need assistance, we can help! Our partners have developed chambers fully compatible with our HD-MEA systems. This enables seamless integration of compartmentalized systems with high-resolution electrophysiology. They bring extensive expertise in designing and fabricating microfluidics devices. The combination of MPS and HD-MEA has already been demonstrated in multiple publications by our users. Whether you want to adopt an existing, validated MPS structure or explore a fully customized design, together with our partner we can accelerate your research. Contact us to discuss our solution for you!
Selected Resources
Sensory-spinal neuron co-culture platform enables analysis of sensory-driven spinal activation

Investigation of the input-output relationship of engineered neural networks using high-density microelectrode arrays

Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks

A magnetically actuated microrobot for targeted neural cell delivery and selective connection of neural networks

PDMS Application Guide
Find important guidelines on PDMS applications and learn how to culture and record from engineered neuronal networks on MaxWell Biosystems' HD-MEA platforms.
MaxOne+ Brochure
Experience MaxOne⁺, the next generation ready-to-use HD-MEA chip with enhanced cell adhesion, superior signal quality, and powerful stimulation for peak performance.