Mapping Neuronal Diversity: Functional Validation of Induced Neurons with HD-MEAs and DeePhys
“Diversity of iN subtypes is evident in their electrical activity patterns, suggesting that they are functionally distinct.”
The human brain, being the absurdly complex system that it is, has been frustrating researchers for centuries. Its complexity starts early on, as stem cells turn into neurons, which then branch into many types and subtypes, all working (or not working) in unison. To build reliable models of the brain, we need to consistently recreate its rich variety of neuronal subtypes in vitro. No one will be surprised then when I say that induced Neurons (iNs) derived from pluripotent stem cells (PSCs) are relevant in the neuroscientific field (understatement of the century), and have become increasingly central to study neural development, disease mechanisms, potential cell replacement therapies... the list goes on (and on and on and on).
Today I want to put a blip on your radar, turning your attention to a paper by Lin et al., published in Science. What they did in this study is incredible to me, and a real breakthrough in the iPSC field: by combining pro-neural transcription factors (TFs) with hundreds of morphogen patterning conditions, they profiled nearly 700,000 cells across 480 conditions, to set up a framework and define strategies for derivation of a diverse set of iN subtypes. Now, if you are thinking “wow, that is a lot of effort, thank you for your incredible service” well, I would agree with you, but it is not over yet.
While single-cell transcriptomics identified these subtypes at the molecular level, the key question was: do these cells also act differently? For neurons, function ultimately comes down to one topic: electrophysiology. To address this important question, the team used the MaxTwo HD-MEA System, which enabled them to record subcellular electrophysiological signatures, single-neuron activity, and network-level dynamics from the same (many) cultures over multiple days (see Figure).

These recording yielded rich functional data regarding their cells, which they turned into detailed insights via DeePhys (Hornauer et al., 2024), an automatic machine learning–driven analysis platform whose developed on MaxWell Biosystems’ recordings. DeePhys automatically extracted and aggregated thousands of electrophysiological features, revealing 11 distinct, single-cell functional phenotypes that - ta-da! - aligned with the morphogen conditions seen at the molecular level! The team also uncovered a clear link between axonal complexity and transcriptional maturity, performing axonal electrical footprint reconstructions which were made possible (and it’s kind of crazy that it’s something that you can just do) by the MaxTwo’s ultra-high spatiotemporal resolution.
What I really like about this study is that you know they were navigating treacherous seas, those of neuronal types and subtypes and the complexity of neuronal development. Nevertheless, they were victorious not by trying to reduce complexity by approximating, but by using the proper tools to explore such complexity in detail. Combining molecular techniques, the high-content and high-resolution of the MaxTwo HD-MEA System, and the advanced/automatic analysis from DeePhys they turned a transcriptomic atlas into a living, functional map of neuronal diversity, where identity and function could be understood side by side. Reads like an epic poem.
If you are interested in exploring how this electrophysiology workflow can be applied to your own research (and yes, it is as cool as it sounds), we have prepared a step-by-step guide that walks through the process of going from MaxWell Biosystems recordings, to spike sorting, to obtaining functional phenotyping insights via: Spike Sorting MaxLab Live Recordings and Phenotyping with SpikeInterface and DeePhys.
Want the full story from the source? Dr. Hsiu-Chuan Lin and Dr. Philipp Hornauer presented a joint webinar on this work. Watch the recording here.
Related
Resources
MaxTwo
Maximize your cell functional assays

MaxLab Live
All-in-one Software
Neuronal Cell Cultures
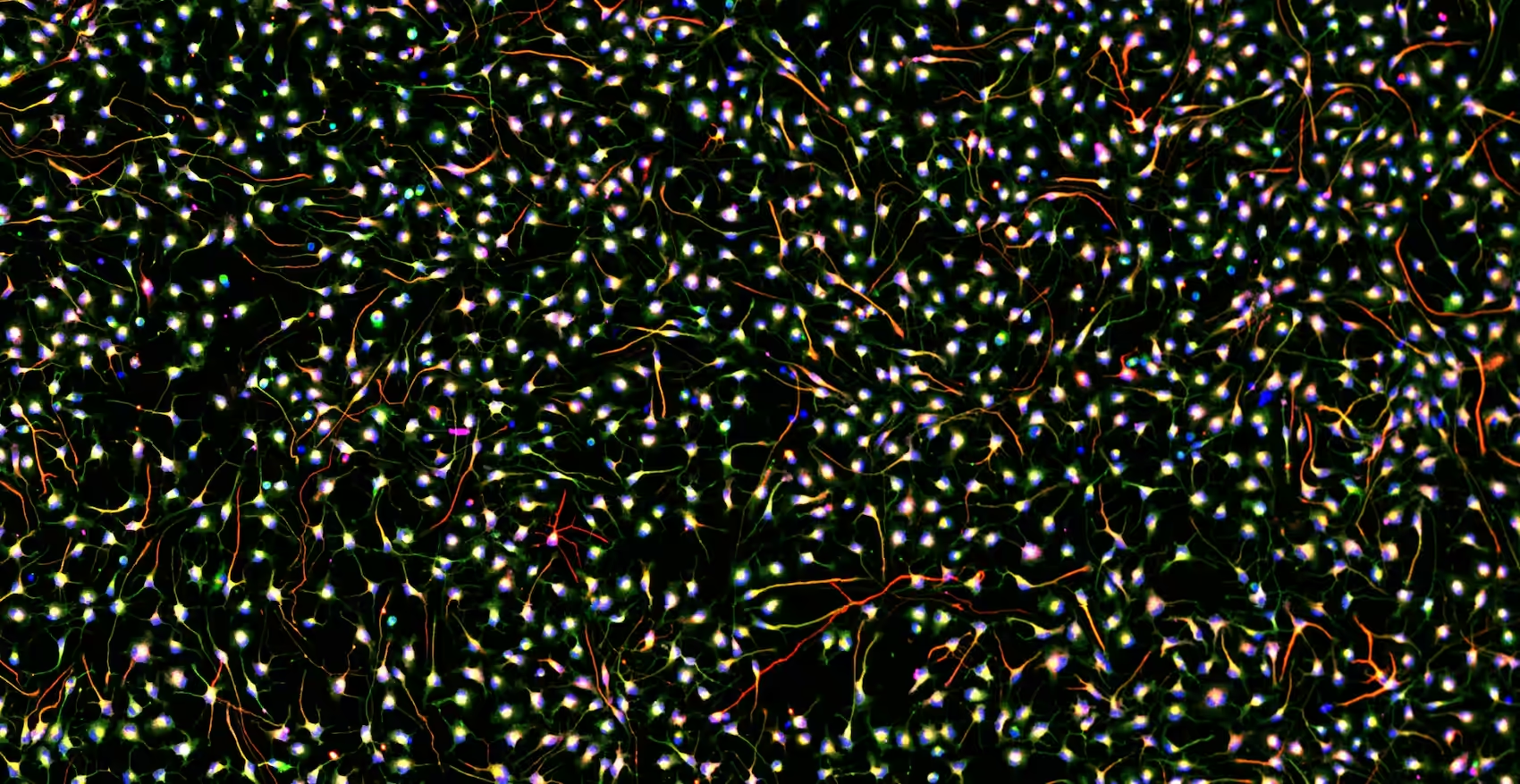
Functional Phenotyping
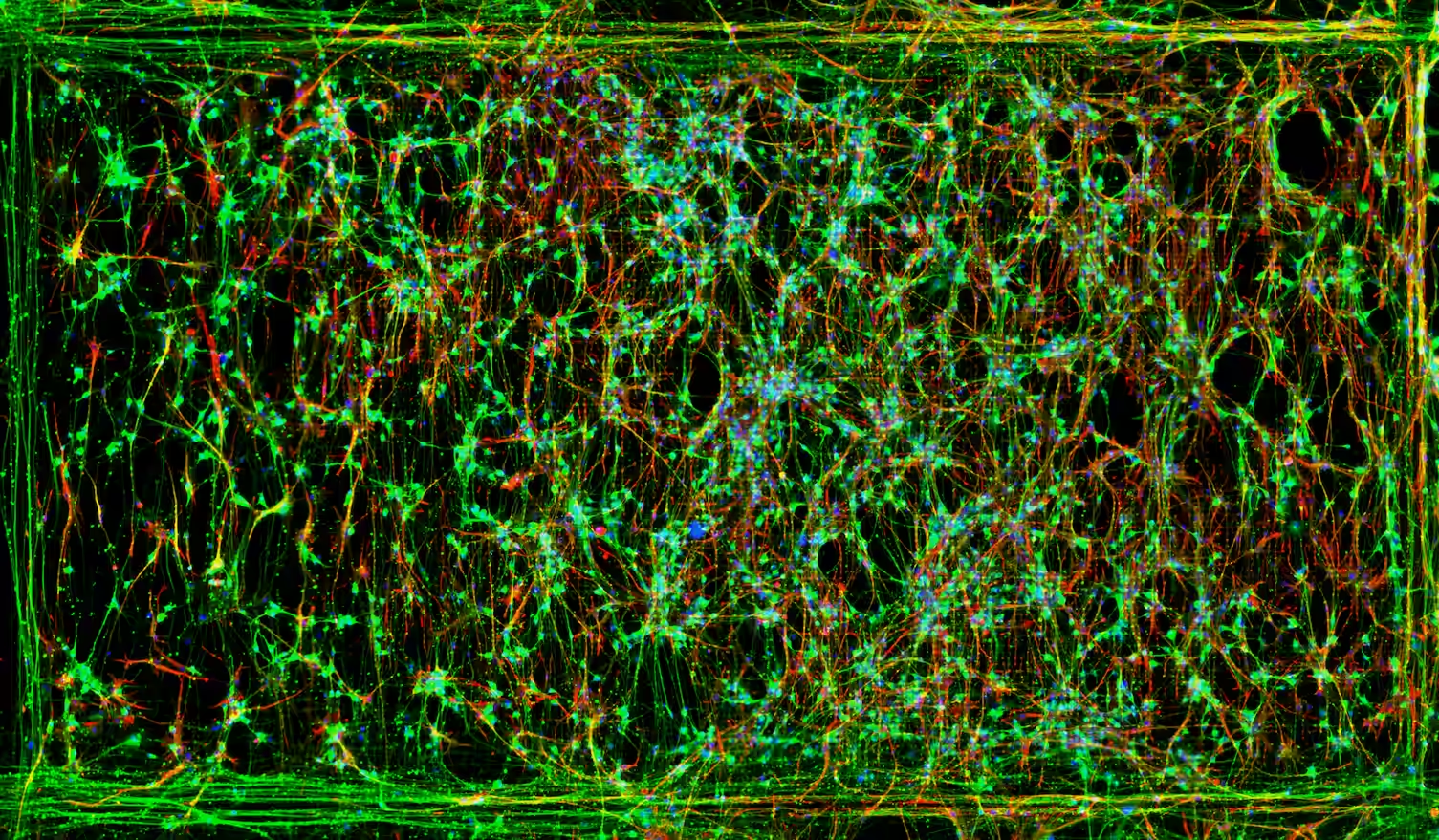
Human neuron subtype programming via single-cell transcriptome-coupled patterning screens

DeePhys: A machine learning–assisted platform for electrophysiological phenotyping of human neuronal networks

