MaxOne High-Density Microelectrode Array (HD-MEA) System
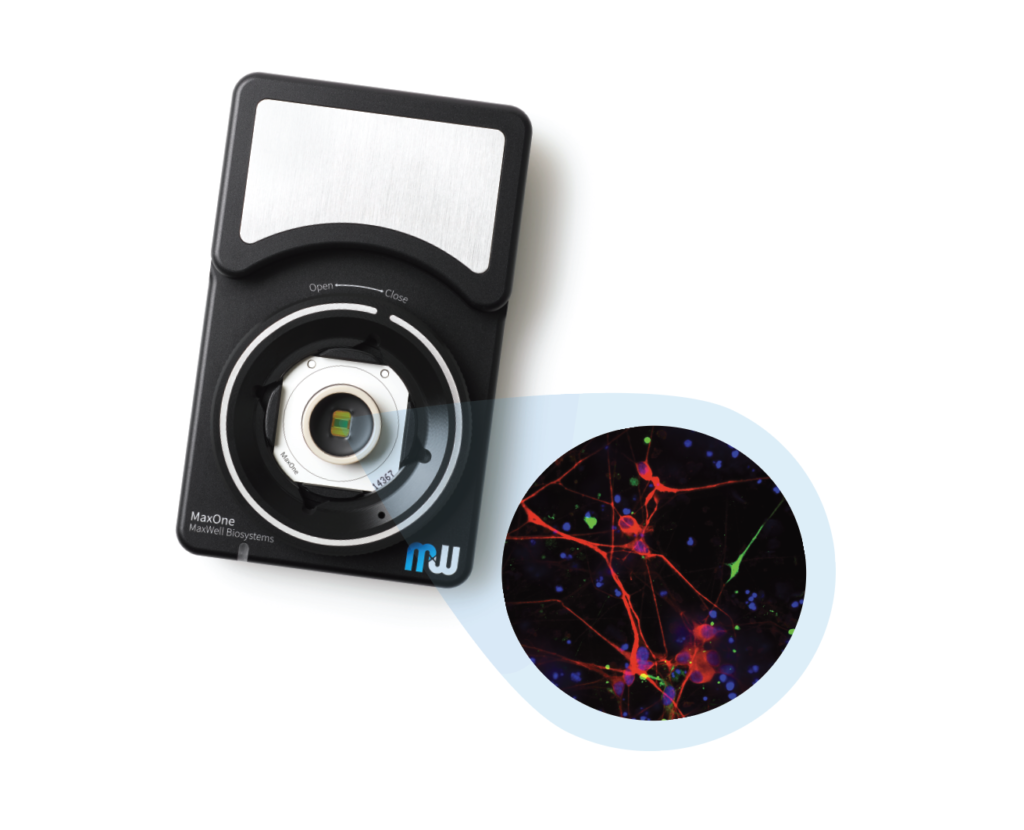

MaxOne – the most powerful electrophysiological platform for recording and stimulating electrogenic cells in vitro. MaxOne is a high-density microelectrode array system in a single-well format.
Key Features


High-resolution and high-quality data while tracking dynamic changes at cellular, subcellular and network levels.

Smallest signals capture (uV) thanks to low-noise recording channels and high electrode density.

Compactly built, MaxOne Recording Unit can be flexibly installed in various environments and is compatible with other devices for both cultured samples and acute tissues.

Optimized recordings strategies to analyze the entire culture at individual neuronal levels, increasing data reproducibility and statistical power.

Cell development, maturation, or compound effects assess by performing longitudinal experiments over the course of days, weeks, and months.


Non-invasive and label-free recordings, eliminating any potential side effects associated with the use of dyes and prolonged exposure to light.
Product Overview
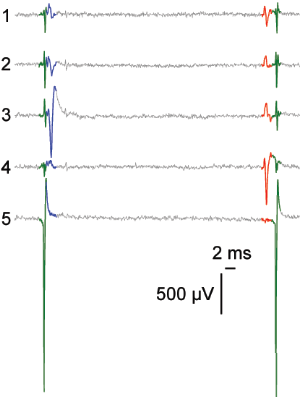
MaxWell Biosystems’s MaxOne is a CMOS-based HD-MEA, an electrical imaging system for neuroscience, drug discovery, and cell assessment applications. MaxOne captures high-quality signals at subcellular resolution from long-term culture experiments and acute tissue experiments (e.g., brain slices, retina).
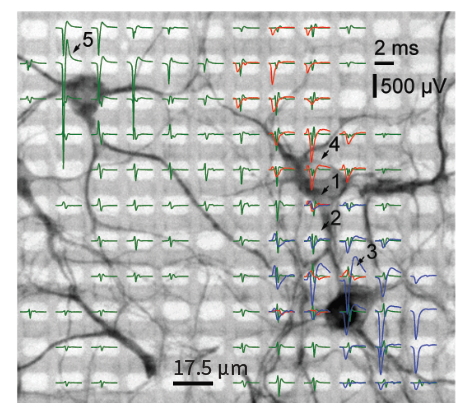

3’625 Electrodes/mm2
Low-Noise Readouts
Flexible Electrical Stimulation

Network level
Cellular level
Subcellular level
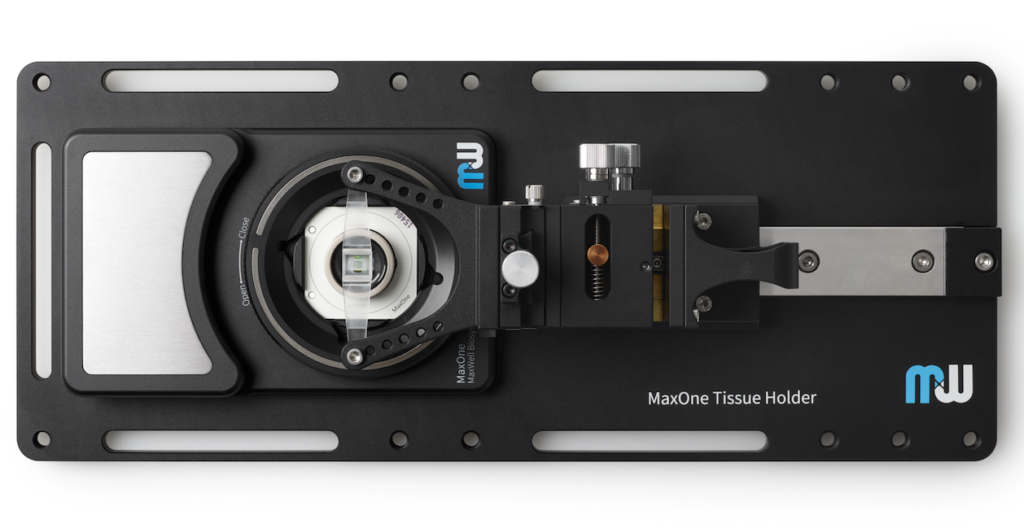

MaxOne HD-MEA system can be flexibly installed in diverse environments. For instance, MaxOne Recording Unit can operate long-term inside cell-culture incubators, and can be integrated with perfusion systems, enabling users to perform acute tissue recordings. Additionally, the system is compatible with other devices, such as being placed under an upright microscope for imaging and microscopy purposes, and integrated with tissue holders for acute tissue experiments.
Learn About MaxOne AccessoriesAll consumable MaxOne Chips share the same core features of our HD-MEA technology but differs in ways that allows them to be more suitable for cultured or acute samples.
Learn About MaxOne Chips

MaxOne System


| MaxOne System Features | ||
|---|---|---|
| Components | Hub Recording Unit | |
| System status indicator | LED | |
| Dimension (L x W x H) | Hub: 18.5 x 28x 9 cm3 Recording Unit: 15 x 9.5 x 2.5 cm3 | |
| Chip compatibility | Small (PSM) Large (PLM) | |
| Incubator friendly | Yes, for the Recording Unit |
This might also interest you



Publications Featuring the MaxOne
  | Fenton, Timothy A; Haouchine, Olivia Y; Hallam, Elizabeth L; Smith, Emily M; Jackson, Kiya C; Rahbarian, Darlene; Canales, Cesar; Adhikari, Anna; Nord, Alexander S; Ben-Shalom, Roy; Silverman, Jill L Hyperexcitability and translational phenotypes in a preclinical mouse model of SYNGAP1-Related Intellectual Disability Journal Article Research Square, 2024. @article{Fenton2024, title = {Hyperexcitability and translational phenotypes in a preclinical mouse model of SYNGAP1-Related Intellectual Disability}, author = {Timothy A Fenton and Olivia Y Haouchine and Elizabeth L Hallam and Emily M Smith and Kiya C Jackson and Darlene Rahbarian and Cesar Canales and Anna Adhikari and Alexander S Nord and Roy Ben-Shalom and Jill L Silverman}, url = {https://www.researchsquare.com/article/rs-4067746/v1}, doi = {10.21203/rs.3.rs-4067746/v1}, year = {2024}, date = {2024-03-19}, journal = {Research Square}, abstract = {Disruption of SYNGAP1 directly causes a genetically identifiable neurodevelopmental disorder (NDD) called SYNGAP1-related intellectual disability (SRID). Without functional SynGAP1 protein, individuals are developmentally delayed and have prominent features of intellectual disability, motor impairments, and epilepsy. Over the past two decades, there have been numerous discoveries indicting the critical role of Syngap1. Several rodent models with a loss of Syngap1 have been engineered identifying precise roles in neuronal structure and function, as well as key biochemical pathways key for synapse integrity. Homozygous loss of Syngap1 is lethal. Heterozygous mutations of Syngap1 result in a broad range of behavioral phenotypes. Our in vivo functional data, using the mouse model from the Huganir laboratory, corroborated earlier reported behaviors including robust hyperactivity and deficits in learning and memory in young adults. In extension, we report impairments in slow wave sleep, a critical component of the domain of sleep. We characterized Syngap1+/- mice by using neurophysiology collected with wireless, telemetric electroencephalography (EEG). Syngap1+/- mice also exhibited elevated spiking events and spike trains, in addition to elevated power, most notably in the delta frequency band. For the first time, we illustrated how primary neurons from Syngap1+/- mice function and display increased network firing activity, greater bursts, and shorter inter-burst intervals between peaks by employing high density microelectrode arrays (HD-MEA). Our reported data bridge in-vitro electrophysiological neuronal activity and function with in vivo neurophysiological brain activity and function. These data elucidate quantitative, translational biomarkers in vivo and in vitro that can be utilized for the development of and efficacy assessment of targeted treatments for SRID.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Disruption of SYNGAP1 directly causes a genetically identifiable neurodevelopmental disorder (NDD) called SYNGAP1-related intellectual disability (SRID). Without functional SynGAP1 protein, individuals are developmentally delayed and have prominent features of intellectual disability, motor impairments, and epilepsy. Over the past two decades, there have been numerous discoveries indicting the critical role of Syngap1. Several rodent models with a loss of Syngap1 have been engineered identifying precise roles in neuronal structure and function, as well as key biochemical pathways key for synapse integrity. Homozygous loss of Syngap1 is lethal. Heterozygous mutations of Syngap1 result in a broad range of behavioral phenotypes. Our in vivo functional data, using the mouse model from the Huganir laboratory, corroborated earlier reported behaviors including robust hyperactivity and deficits in learning and memory in young adults. In extension, we report impairments in slow wave sleep, a critical component of the domain of sleep. We characterized Syngap1+/- mice by using neurophysiology collected with wireless, telemetric electroencephalography (EEG). Syngap1+/- mice also exhibited elevated spiking events and spike trains, in addition to elevated power, most notably in the delta frequency band. For the first time, we illustrated how primary neurons from Syngap1+/- mice function and display increased network firing activity, greater bursts, and shorter inter-burst intervals between peaks by employing high density microelectrode arrays (HD-MEA). Our reported data bridge in-vitro electrophysiological neuronal activity and function with in vivo neurophysiological brain activity and function. These data elucidate quantitative, translational biomarkers in vivo and in vitro that can be utilized for the development of and efficacy assessment of targeted treatments for SRID. |
  | Voitiuk, Kateryna; Seiler, Spencer T; de Melo, Mirella Pessoa; Geng, Jinghui; Hernandez, Sebastian; Schweiger, Hunter E; Sevetson, Jess L; Parks, David F; Robbins, Ash; Torres-Montoya, Sebastian; Ehrlich, Drew; Elliott, Matthew A T; Sharf, Tal; Haussler, David; Mostajo-Radji, Mohammed A; Salama, Sofie R; Teodorescu, Mircea A feedback-driven IoT microfluidic, electrophysiology, and imaging platform for brain organoid studies Journal Article bioRxiv, 2024. @article{Voitiuk2024, title = {A feedback-driven IoT microfluidic, electrophysiology, and imaging platform for brain organoid studies}, author = {Kateryna Voitiuk and Spencer T. Seiler and Mirella Pessoa de Melo and Jinghui Geng and Sebastian Hernandez and Hunter E. Schweiger and Jess L. Sevetson and David F. Parks and Ash Robbins and Sebastian Torres-Montoya and Drew Ehrlich and Matthew A.T. Elliott and Tal Sharf and David Haussler and Mohammed A. Mostajo-Radji and Sofie R. Salama and Mircea Teodorescu}, url = {http://biorxiv.org/lookup/doi/10.1101/2024.03.15.585237}, doi = {10.1101/2024.03.15.585237}, year = {2024}, date = {2024-03-17}, journal = {bioRxiv}, abstract = {The analysis of tissue cultures, particularly brain organoids, takes a high degree of coordination, measurement, and monitoring. We have developed an automated research platform enabling independent devices to achieve collaborative objectives for feedback-driven cell culture studies. Unified by an Internet of Things (IoT) architecture, our approach enables continuous, communicative interactions among various sensing and actuation devices, achieving precisely timed control of in vitro biological experiments. The framework integrates microfluidics, electrophysiology, and imaging devices to maintain cerebral cortex organoids and monitor their neuronal activity. The organoids are cultured in custom, 3D-printed chambers attached to commercial microelectrode arrays for electrophysiology monitoring. Periodic feeding is achieved using programmable microfluidic pumps. We developed computer vision fluid volume estimations of aspirated media, achieving high accuracy, and used feedback to rectify deviations in microfluidic perfusion during media feeding/aspiration cycles. We validated the system with a 7-day study of mouse cerebral cortex organoids, comparing manual and automated protocols. The automated experimental samples maintained robust neural activity throughout the experiment, comparable with the control samples. The automated system enabled hourly electrophysiology recordings that revealed dramatic temporal changes in neuron firing rates not observed in once-a-day recordings. One-Sentence Summary: An IoT laboratory robotics system that enables touch-free feeding, imaging, and electrophysiology of brain organoids.}, keywords = {}, pubstate = {published}, tppubtype = {article} } The analysis of tissue cultures, particularly brain organoids, takes a high degree of coordination, measurement, and monitoring. We have developed an automated research platform enabling independent devices to achieve collaborative objectives for feedback-driven cell culture studies. Unified by an Internet of Things (IoT) architecture, our approach enables continuous, communicative interactions among various sensing and actuation devices, achieving precisely timed control of in vitro biological experiments. The framework integrates microfluidics, electrophysiology, and imaging devices to maintain cerebral cortex organoids and monitor their neuronal activity. The organoids are cultured in custom, 3D-printed chambers attached to commercial microelectrode arrays for electrophysiology monitoring. Periodic feeding is achieved using programmable microfluidic pumps. We developed computer vision fluid volume estimations of aspirated media, achieving high accuracy, and used feedback to rectify deviations in microfluidic perfusion during media feeding/aspiration cycles. We validated the system with a 7-day study of mouse cerebral cortex organoids, comparing manual and automated protocols. The automated experimental samples maintained robust neural activity throughout the experiment, comparable with the control samples. The automated system enabled hourly electrophysiology recordings that revealed dramatic temporal changes in neuron firing rates not observed in once-a-day recordings. One-Sentence Summary: An IoT laboratory robotics system that enables touch-free feeding, imaging, and electrophysiology of brain organoids. |
  | Kasuba, Krishna Chaitanya; Buccino, Alessio Paolo; Bartram, Julian; Gaub, Benjamin M; Fauser, Felix J; Ronchi, Silvia; Kumar, Sreedhar Saseendran; Geissler, Sydney; Nava, Michele M; Hierlemann, Andreas; Müller, Daniel J Nature Nanotechnology, 2024. @article{Kasuba2024, title = {Mechanical stimulation and electrophysiological monitoring at subcellular resolution reveals differential mechanosensation of neurons within networks}, author = {Krishna Chaitanya Kasuba and Alessio Paolo Buccino and Julian Bartram and Benjamin M. Gaub and Felix J. Fauser and Silvia Ronchi and Sreedhar Saseendran Kumar and Sydney Geissler and Michele M. Nava and Andreas Hierlemann and Daniel J. Müller }, url = {https://www.nature.com/articles/s41565-024-01609-1}, doi = {10.1038/s41565-024-01609-1}, year = {2024}, date = {2024-02-20}, journal = {Nature Nanotechnology}, abstract = {A growing consensus that the brain is a mechanosensitive organ is driving the need for tools that mechanically stimulate and simultaneously record the electrophysiological response of neurons within neuronal networks. Here we introduce a synchronized combination of atomic force microscopy, high-density microelectrode array and fluorescence microscopy to monitor neuronal networks and to mechanically characterize and stimulate individual neurons at piconewton force sensitivity and nanometre precision while monitoring their electrophysiological activity at subcellular spatial and millisecond temporal resolution. No correlation is found between mechanical stiffness and electrophysiological activity of neuronal compartments. Furthermore, spontaneously active neurons show exceptional functional resilience to static mechanical compression of their soma. However, application of fast transient (∼500 ms) mechanical stimuli to the neuronal soma can evoke action potentials, which depend on the anchoring of neuronal membrane and actin cytoskeleton. Neurons show higher responsivity, including bursts of action potentials, to slower transient mechanical stimuli (∼60 s). Moreover, transient and repetitive application of the same compression modulates the neuronal firing rate. Seemingly, neuronal networks can differentiate and respond to specific characteristics of mechanical stimulation. Ultimately, the developed multiparametric tool opens the door to explore manifold nanomechanobiological responses of neuronal systems and new ways of mechanical control.}, keywords = {}, pubstate = {published}, tppubtype = {article} } A growing consensus that the brain is a mechanosensitive organ is driving the need for tools that mechanically stimulate and simultaneously record the electrophysiological response of neurons within neuronal networks. Here we introduce a synchronized combination of atomic force microscopy, high-density microelectrode array and fluorescence microscopy to monitor neuronal networks and to mechanically characterize and stimulate individual neurons at piconewton force sensitivity and nanometre precision while monitoring their electrophysiological activity at subcellular spatial and millisecond temporal resolution. No correlation is found between mechanical stiffness and electrophysiological activity of neuronal compartments. Furthermore, spontaneously active neurons show exceptional functional resilience to static mechanical compression of their soma. However, application of fast transient (∼500 ms) mechanical stimuli to the neuronal soma can evoke action potentials, which depend on the anchoring of neuronal membrane and actin cytoskeleton. Neurons show higher responsivity, including bursts of action potentials, to slower transient mechanical stimuli (∼60 s). Moreover, transient and repetitive application of the same compression modulates the neuronal firing rate. Seemingly, neuronal networks can differentiate and respond to specific characteristics of mechanical stimulation. Ultimately, the developed multiparametric tool opens the door to explore manifold nanomechanobiological responses of neuronal systems and new ways of mechanical control. |
  | Hruska-Plochan, Marian; Wiersma, Vera I; Betz, Katharina M; Mallona, Izaskun; Ronchi, Silvia; Maniecka, Zuzanna; Hock, Eva-Maria; Tantardini, Elena; Laferriere, Florent; Sahadevan, Sonu; Hoop, Vanessa; Delvendahl, Igor; Pérez-Berlanga, Manuela; Gatta, Beatrice; Panatta, Martina; van der Bourg, Alexander; Bohaciakova, Dasa; Sharma, Puneet; Vos, Laura De; Frontzek, Karl; Aguzzi, Adriano; Lashley, Tammaryn; Robinson, Mark D; Karayannis, Theofanis; Mueller, Martin; Hierlemann, Andreas; Polymenidou, Magdalini A model of human neural networks reveals NPTX2 pathology in ALS and FTLD Journal Article Nature, 2024. @article{Hruska-Plochan2024, title = {A model of human neural networks reveals NPTX2 pathology in ALS and FTLD}, author = {Marian Hruska-Plochan and Vera I. Wiersma and Katharina M. Betz and Izaskun Mallona and Silvia Ronchi and Zuzanna Maniecka and Eva-Maria Hock and Elena Tantardini and Florent Laferriere and Sonu Sahadevan and Vanessa Hoop and Igor Delvendahl and Manuela Pérez-Berlanga and Beatrice Gatta and Martina Panatta and Alexander van der Bourg and Dasa Bohaciakova and Puneet Sharma and Laura De Vos and Karl Frontzek and Adriano Aguzzi and Tammaryn Lashley and Mark D. Robinson and Theofanis Karayannis and Martin Mueller and Andreas Hierlemann and Magdalini Polymenidou }, url = {https://www.nature.com/articles/s41586-024-07042-7}, doi = {10.1038/s41586-024-07042-7}, year = {2024}, date = {2024-02-14}, journal = {Nature}, abstract = {Human cellular models of neurodegeneration require reproducibility and longevity, which is necessary for simulating age-dependent diseases. Such systems are particularly needed for TDP-43 proteinopathies1, which involve human-specific mechanisms that cannot be directly studied in animal models. Here, to explore the emergence and consequences of TDP-43 pathologies, we generated induced pluripotent stem cell-derived, colony morphology neural stem cells (iCoMoNSCs) via manual selection of neural precursors. Single-cell transcriptomics and comparison to independent neural stem cells showed that iCoMoNSCs are uniquely homogenous and self-renewing. Differentiated iCoMoNSCs formed a self-organized multicellular system consisting of synaptically connected and electrophysiologically active neurons, which matured into long-lived functional networks (which we designate iNets). Neuronal and glial maturation in iNets was similar to that of cortical organoids. Overexpression of wild-type TDP-43 in a minority of neurons within iNets led to progressive fragmentation and aggregation of the protein, resulting in a partial loss of function and neurotoxicity. Single-cell transcriptomics revealed a novel set of misregulated RNA targets in TDP-43-overexpressing neurons and in patients with TDP-43 proteinopathies exhibiting a loss of nuclear TDP-43. The strongest misregulated target encoded the synaptic protein NPTX2, the levels of which are controlled by TDP-43 binding on its 3′ untranslated region. When NPTX2 was overexpressed in iNets, it exhibited neurotoxicity, whereas correcting NPTX2 misregulation partially rescued neurons from TDP-43-induced neurodegeneration. Notably, NPTX2 was consistently misaccumulated in neurons from patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration with TDP-43 pathology. Our work directly links TDP-43 misregulation and NPTX2 accumulation, thereby revealing a TDP-43-dependent pathway of neurotoxicity.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Human cellular models of neurodegeneration require reproducibility and longevity, which is necessary for simulating age-dependent diseases. Such systems are particularly needed for TDP-43 proteinopathies1, which involve human-specific mechanisms that cannot be directly studied in animal models. Here, to explore the emergence and consequences of TDP-43 pathologies, we generated induced pluripotent stem cell-derived, colony morphology neural stem cells (iCoMoNSCs) via manual selection of neural precursors. Single-cell transcriptomics and comparison to independent neural stem cells showed that iCoMoNSCs are uniquely homogenous and self-renewing. Differentiated iCoMoNSCs formed a self-organized multicellular system consisting of synaptically connected and electrophysiologically active neurons, which matured into long-lived functional networks (which we designate iNets). Neuronal and glial maturation in iNets was similar to that of cortical organoids. Overexpression of wild-type TDP-43 in a minority of neurons within iNets led to progressive fragmentation and aggregation of the protein, resulting in a partial loss of function and neurotoxicity. Single-cell transcriptomics revealed a novel set of misregulated RNA targets in TDP-43-overexpressing neurons and in patients with TDP-43 proteinopathies exhibiting a loss of nuclear TDP-43. The strongest misregulated target encoded the synaptic protein NPTX2, the levels of which are controlled by TDP-43 binding on its 3′ untranslated region. When NPTX2 was overexpressed in iNets, it exhibited neurotoxicity, whereas correcting NPTX2 misregulation partially rescued neurons from TDP-43-induced neurodegeneration. Notably, NPTX2 was consistently misaccumulated in neurons from patients with amyotrophic lateral sclerosis and frontotemporal lobar degeneration with TDP-43 pathology. Our work directly links TDP-43 misregulation and NPTX2 accumulation, thereby revealing a TDP-43-dependent pathway of neurotoxicity. |
  | Hornauer, Philipp; Prack, Gustavo; Anastasi, Nadia; Ronchi, Silvia; Kim, Taehoon; Donner, Christian; Fiscella, Michele; Borgwardt, Karsten; Taylor, Verdon; Jagasia, Ravi; Roqueiro, Damian; Hierlemann, Andreas; Schröter, Manuel DeePhys: A machine learning–assisted platform for electrophysiological phenotyping of human neuronal networks Journal Article Stem Cell Reports, 2024. @article{Hornauer2024, title = {DeePhys: A machine learning–assisted platform for electrophysiological phenotyping of human neuronal networks}, author = {Philipp Hornauer and Gustavo Prack and Nadia Anastasi and Silvia Ronchi and Taehoon Kim and Christian Donner and Michele Fiscella and Karsten Borgwardt and Verdon Taylor and Ravi Jagasia and Damian Roqueiro and Andreas Hierlemann and Manuel Schröter}, url = {https://www.sciencedirect.com/science/article/pii/S2213671123005015}, doi = {10.1016/j.stemcr.2023.12.008}, year = {2024}, date = {2024-01-25}, journal = {Stem Cell Reports}, abstract = {Reproducible functional assays to study in vitro neuronal networks represent an important cornerstone in the quest to develop physiologically relevant cellular models of human diseases. Here, we introduce DeePhys, a MATLAB-based analysis tool for data-driven functional phenotyping of in vitro neuronal cultures recorded by high-density microelectrode arrays. DeePhys is a modular workflow that offers a range of techniques to extract features from spike-sorted data, allowing for the examination of functional phenotypes both at the individual cell and network levels, as well as across development. In addition, DeePhys incorporates the capability to integrate novel features and to use machine-learning-assisted approaches, which facilitates a comprehensive evaluation of pharmacological interventions. To illustrate its practical application, we apply DeePhys to human induced pluripotent stem cell–derived dopaminergic neurons obtained from both patients and healthy individuals and showcase how DeePhys enables phenotypic screenings.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Reproducible functional assays to study in vitro neuronal networks represent an important cornerstone in the quest to develop physiologically relevant cellular models of human diseases. Here, we introduce DeePhys, a MATLAB-based analysis tool for data-driven functional phenotyping of in vitro neuronal cultures recorded by high-density microelectrode arrays. DeePhys is a modular workflow that offers a range of techniques to extract features from spike-sorted data, allowing for the examination of functional phenotypes both at the individual cell and network levels, as well as across development. In addition, DeePhys incorporates the capability to integrate novel features and to use machine-learning-assisted approaches, which facilitates a comprehensive evaluation of pharmacological interventions. To illustrate its practical application, we apply DeePhys to human induced pluripotent stem cell–derived dopaminergic neurons obtained from both patients and healthy individuals and showcase how DeePhys enables phenotypic screenings. |
  | van der Molen, Tjitse; Spaeth, Alex; Chini, Mattia; Bartram, Julian; Dendukuri, Aditya; Zhang, Zongren; Bhaskaran-Nair, Kiran; Blauvelt, Lon J; Petzold, Linda R; Hansma, Paul K; Teodorescu, Mircea; Hierlemann, Andreas; Hengen, Keith B; Hanganu-Opatz, Ileana L; Kosik, Kenneth S; Sharf, Tal Protosequences in human cortical organoids model intrinsic states in the developing cortex Journal Article bioRxiv, 2023. @article{Molen2023, title = {Protosequences in human cortical organoids model intrinsic states in the developing cortex}, author = {Tjitse van der Molen and Alex Spaeth and Mattia Chini and Julian Bartram and Aditya Dendukuri and Zongren Zhang and Kiran Bhaskaran-Nair and Lon J. Blauvelt and Linda R. Petzold and Paul K. Hansma and Mircea Teodorescu and Andreas Hierlemann and Keith B. Hengen and Ileana L. Hanganu-Opatz and Kenneth S. Kosik and Tal Sharf}, url = {https://www.biorxiv.org/content/10.1101/2023.12.29.573646v1}, doi = {10.1101/2023.12.29.573646}, year = {2023}, date = {2023-12-30}, journal = {bioRxiv}, abstract = {Neuronal firing sequences are thought to be the basic building blocks of neural coding and information broadcasting within the brain. However, when sequences emerge during neurodevelopment remains unknown. We demonstrate that structured firing sequences are present in spontaneous activity of human brain organoids and ex vivo neonatal brain slices from the murine somatosensory cortex. We observed a balance between temporally rigid and flexible firing patterns that are emergent phenomena in human brain organoids and early postnatal murine somatosensory cortex, but not in primary dissociated cortical cultures. Our findings suggest that temporal sequences do not arise in an experience-dependent manner, but are rather constrained by an innate preconfigured architecture established during neurogenesis. These findings highlight the potential for brain organoids to further explore how exogenous inputs can be used to refine neuronal circuits and enable new studies into the genetic mechanisms that govern assembly of functional circuitry during early human brain development.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Neuronal firing sequences are thought to be the basic building blocks of neural coding and information broadcasting within the brain. However, when sequences emerge during neurodevelopment remains unknown. We demonstrate that structured firing sequences are present in spontaneous activity of human brain organoids and ex vivo neonatal brain slices from the murine somatosensory cortex. We observed a balance between temporally rigid and flexible firing patterns that are emergent phenomena in human brain organoids and early postnatal murine somatosensory cortex, but not in primary dissociated cortical cultures. Our findings suggest that temporal sequences do not arise in an experience-dependent manner, but are rather constrained by an innate preconfigured architecture established during neurogenesis. These findings highlight the potential for brain organoids to further explore how exogenous inputs can be used to refine neuronal circuits and enable new studies into the genetic mechanisms that govern assembly of functional circuitry during early human brain development. |
  | Cai, Hongwei; Ao, Zheng; Tian, Chunhui; Wu, Zhuhao; Liu, Hongcheng; Tchieu, Jason; Gu, Mingxia; Mackie, Ken; Guo, Feng Brain organoid reservoir computing for artificial intelligence Journal Article Nature Electronics, 2023. @article{Cai2023b, title = {Brain organoid reservoir computing for artificial intelligence}, author = {Hongwei Cai and Zheng Ao and Chunhui Tian and Zhuhao Wu and Hongcheng Liu and Jason Tchieu and Mingxia Gu and Ken Mackie and Feng Guo }, url = {https://www.nature.com/articles/s41928-023-01069-w}, doi = {10.1038/s41928-023-01069-w}, year = {2023}, date = {2023-12-11}, journal = {Nature Electronics}, abstract = {Brain-inspired computing hardware aims to emulate the structure and working principles of the brain and could be used to address current limitations in artificial intelligence technologies. However, brain-inspired silicon chips are still limited in their ability to fully mimic brain function as most examples are built on digital electronic principles. Here we report an artificial intelligence hardware approach that uses adaptive reservoir computation of biological neural networks in a brain organoid. In this approach—which is termed Brainoware—computation is performed by sending and receiving information from the brain organoid using a high-density multielectrode array. By applying spatiotemporal electrical stimulation, nonlinear dynamics and fading memory properties are achieved, as well as unsupervised learning from training data by reshaping the organoid functional connectivity. We illustrate the practical potential of this technique by using it for speech recognition and nonlinear equation prediction in a reservoir computing framework.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Brain-inspired computing hardware aims to emulate the structure and working principles of the brain and could be used to address current limitations in artificial intelligence technologies. However, brain-inspired silicon chips are still limited in their ability to fully mimic brain function as most examples are built on digital electronic principles. Here we report an artificial intelligence hardware approach that uses adaptive reservoir computation of biological neural networks in a brain organoid. In this approach—which is termed Brainoware—computation is performed by sending and receiving information from the brain organoid using a high-density multielectrode array. By applying spatiotemporal electrical stimulation, nonlinear dynamics and fading memory properties are achieved, as well as unsupervised learning from training data by reshaping the organoid functional connectivity. We illustrate the practical potential of this technique by using it for speech recognition and nonlinear equation prediction in a reservoir computing framework. |
  | Elliott, Matthew A T; Schweiger, Hunter E; Robbins, Ash; Vera-Choqqueccota, Samira; Ehrlich, Drew; Hernandez, Sebastian; Voitiuk, Kateryna; Geng, Jinghui; Sevetson, Jess L; Core, Cordero; Rosen, Yohei M; Teodorescu, Mircea; Wagner, Nico O; Haussler, David; Mostajo-Radji, Mohammed A Internet-Connected Cortical Organoids for Project-Based Stem Cell and Neuroscience Education Journal Article eNeuro, 2023. @article{Elliott2023, title = {Internet-Connected Cortical Organoids for Project-Based Stem Cell and Neuroscience Education}, author = {Matthew A. T. Elliott and Hunter E. Schweiger and Ash Robbins and Samira Vera-Choqqueccota and Drew Ehrlich and Sebastian Hernandez and Kateryna Voitiuk and Jinghui Geng and Jess L. Sevetson and Cordero Core and Yohei M. Rosen and Mircea Teodorescu and Nico O. Wagner and David Haussler and Mohammed A. Mostajo-Radji}, url = {https://www.eneuro.org/lookup/doi/10.1523/ENEURO.0308-23.2023}, doi = {10.1523/ENEURO.0308-23.2023}, year = {2023}, date = {2023-11-28}, journal = {eNeuro}, abstract = {The introduction of Internet-connected technologies to the classroom has the potential to revolutionize STEM education by allowing students to perform experiments in complex models that are unattainable in traditional teaching laboratories. By connecting laboratory equipment to the cloud, we introduce students to experimentation in pluripotent stem cell (PSC)-derived cortical organoids in two different settings: using microscopy to monitor organoid growth in an introductory tissue culture course and using high-density (HD) multielectrode arrays (MEAs) to perform neuronal stimulation and recording in an advanced neuroscience mathematics course. We demonstrate that this approach develops interest in stem cell and neuroscience in the students of both courses. All together, we propose cloud technologies as an effective and scalable approach for complex project-based university training.}, keywords = {}, pubstate = {published}, tppubtype = {article} } The introduction of Internet-connected technologies to the classroom has the potential to revolutionize STEM education by allowing students to perform experiments in complex models that are unattainable in traditional teaching laboratories. By connecting laboratory equipment to the cloud, we introduce students to experimentation in pluripotent stem cell (PSC)-derived cortical organoids in two different settings: using microscopy to monitor organoid growth in an introductory tissue culture course and using high-density (HD) multielectrode arrays (MEAs) to perform neuronal stimulation and recording in an advanced neuroscience mathematics course. We demonstrate that this approach develops interest in stem cell and neuroscience in the students of both courses. All together, we propose cloud technologies as an effective and scalable approach for complex project-based university training. |
  | Kobayashi, Toki; Shimba, Kenta; Kotani, Kiyoshi; Jimbo, Yasuhiko Measuring and Inducing the Plasticity of Single-Neuron Scale at Multiple Points Conference ISSS Xplore, 2023. @conference{Kobayashi2023, title = {Measuring and Inducing the Plasticity of Single-Neuron Scale at Multiple Points}, author = {Toki Kobayashi and Kenta Shimba and Kiyoshi Kotani and Yasuhiko Jimbo}, url = {https://ieeexplore.ieee.org/abstract/document/10322076}, doi = {10.1109/BMEiCON60347.2023.10322076}, year = {2023}, date = {2023-11-22}, journal = {IEEE Xplore}, publisher = {ISSS Xplore}, abstract = {Information processing in the brain is supported by the plasticity of the connections of neurons in biological neuronal networks. There is a gap between our understanding on plasticity of individual connections and that of the neuronal networks. Here, we aimed to induce and measure plasticity at the connections of individual neurons and at the network level. We achieved inducing plasticity with a combination of single cell stimulation by optogenetics and high-resolution extracellular potentials recording by high-density microelectrode array. Spike timing plasticity in a single neuron was successfully measured. Although we also tried to induce synaptic potentiation by tetanus stimulation, no significant change was observed. In the future, we will investigate how the plasticity of individual connections changes dynamics of entire network.}, keywords = {}, pubstate = {published}, tppubtype = {conference} } Information processing in the brain is supported by the plasticity of the connections of neurons in biological neuronal networks. There is a gap between our understanding on plasticity of individual connections and that of the neuronal networks. Here, we aimed to induce and measure plasticity at the connections of individual neurons and at the network level. We achieved inducing plasticity with a combination of single cell stimulation by optogenetics and high-resolution extracellular potentials recording by high-density microelectrode array. Spike timing plasticity in a single neuron was successfully measured. Although we also tried to induce synaptic potentiation by tetanus stimulation, no significant change was observed. In the future, we will investigate how the plasticity of individual connections changes dynamics of entire network. |
  | Tamatani, Chie; Shimba, Kenta; andYasuhiko Jimbo, Kiyoshi Kotani Activity- and Spatial-dependent Variations in Axonal Conduction Recorded from Microtunnel Electrodes Conference IEEE Xplore, 2023. @conference{Tamatani2023, title = {Activity- and Spatial-dependent Variations in Axonal Conduction Recorded from Microtunnel Electrodes}, author = {Chie Tamatani and Kenta Shimba and Kiyoshi Kotani andYasuhiko Jimbo }, url = {https://ieeexplore.ieee.org/abstract/document/10321980}, doi = {10.1109/BMEiCON60347.2023.10321980}, year = {2023}, date = {2023-11-22}, publisher = {IEEE Xplore}, abstract = {Axons are not simple transmission cables in neuronal networks, but they are directly involved in information processing. We developed a novel culture device that aims to assess axonal conduction properties. The microdevice consists of microtunnels directly fabricated on high-density microelectrode array. Neuron growth and axon isolation were confirmed by immunofluorescent staining and neural activity recording. Several dozens of electrodes were detected from each tunnel and signals had enough S/N ratio to handle each spike event. Activity- and spatial-dependent variations in conduction velocity were observed. These results suggest that our microdevice is feasible to study axonal information processing.}, keywords = {}, pubstate = {published}, tppubtype = {conference} } Axons are not simple transmission cables in neuronal networks, but they are directly involved in information processing. We developed a novel culture device that aims to assess axonal conduction properties. The microdevice consists of microtunnels directly fabricated on high-density microelectrode array. Neuron growth and axon isolation were confirmed by immunofluorescent staining and neural activity recording. Several dozens of electrodes were detected from each tunnel and signals had enough S/N ratio to handle each spike event. Activity- and spatial-dependent variations in conduction velocity were observed. These results suggest that our microdevice is feasible to study axonal information processing. |
  | Duru, Jens; Rüfenacht, Arielle; Löhle, Josephine; Pozzi, Marcello; Forró, Csaba; Ledermann, Linus; Bernardi, Aeneas; Matter, Michael; Renia, André; Simona, Benjamin; Tringides, Christina M; Bernhard, Stéphane; Ihle, Stephan J; Hengsteler, Julian; Maurer, Benedikt; Zhanga, Xinyu; Nakatsuka, Nako Driving electrochemical reactions at the microscale using CMOS microelectrode arrays Journal Article Lab on a Chip, 2023, ISSN: 1473-0189. @article{Duru2023c, title = {Driving electrochemical reactions at the microscale using CMOS microelectrode arrays}, author = {Jens Duru and Arielle Rüfenacht and Josephine Löhle and Marcello Pozzi and Csaba Forró and Linus Ledermann and Aeneas Bernardi and Michael Matter and André Renia and Benjamin Simona and Christina M. Tringides and Stéphane Bernhard and Stephan J. Ihle and Julian Hengsteler and Benedikt Maurer and Xinyu Zhanga and Nako Nakatsuka}, url = {https://pubs.rsc.org/en/content/articlelanding/2023/lc/d3lc00630a}, doi = {10.1039/D3LC00630A}, issn = {1473-0189}, year = {2023}, date = {2023-10-30}, journal = {Lab on a Chip}, abstract = {Precise control of pH values at electrode interfaces enables the systematic investigation of pH-dependent processes by electrochemical means. In this work, we employed high-density complementary metal-oxide-semiconductor (CMOS) microelectrode arrays (MEAs) as miniaturized systems to induce and confine electrochemical reactions in areas corresponding to the pitch of single electrodes (17.5 μm). First, we present a strategy for generating localized pH patterns on the surface of the CMOS MEA with unprecedented spatial resolution. Leveraging the versatile routing capabilities of the switch matrix beneath the CMOS MEA, we created arbitrary combinations of anodic and cathodic electrodes and hence pH patterns. Moreover, we utilized the system to produce polymeric surface patterns by additive and subtractive methods. For additive patterning, we controlled the in situ formation of polydopamine at the microelectrode surface through oxidation of free dopamine above a threshold pH > 8.5. For subtractive patterning, we removed cell-adhesive poly-L-lysine from the electrode surface and backfilled the voids with antifouling polymers. Such polymers were chosen to provide a proof-of-concept application of controlling neuronal growth via electrochemically-induced patterns on the CMOS MEA surface. Importantly, our platform is compatible with commercially available high-density MEAs and requires no custom equipment, rendering the findings generalizable and accessible.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Precise control of pH values at electrode interfaces enables the systematic investigation of pH-dependent processes by electrochemical means. In this work, we employed high-density complementary metal-oxide-semiconductor (CMOS) microelectrode arrays (MEAs) as miniaturized systems to induce and confine electrochemical reactions in areas corresponding to the pitch of single electrodes (17.5 μm). First, we present a strategy for generating localized pH patterns on the surface of the CMOS MEA with unprecedented spatial resolution. Leveraging the versatile routing capabilities of the switch matrix beneath the CMOS MEA, we created arbitrary combinations of anodic and cathodic electrodes and hence pH patterns. Moreover, we utilized the system to produce polymeric surface patterns by additive and subtractive methods. For additive patterning, we controlled the in situ formation of polydopamine at the microelectrode surface through oxidation of free dopamine above a threshold pH > 8.5. For subtractive patterning, we removed cell-adhesive poly-L-lysine from the electrode surface and backfilled the voids with antifouling polymers. Such polymers were chosen to provide a proof-of-concept application of controlling neuronal growth via electrochemically-induced patterns on the CMOS MEA surface. Importantly, our platform is compatible with commercially available high-density MEAs and requires no custom equipment, rendering the findings generalizable and accessible. |
  | Metto, Abigael C; Telgkamp, Petra; McLane-Svoboda, Autumn K; Gilad, Assaf A; Pelled, Galit Brain Research, 2023. @article{Metto2023, title = {Closed-loop neurostimulation via expression of magnetogenetics-sensitive protein in inhibitory neurons leads to reduction of seizure activity in a rat model of epilepsy}, author = {Abigael C. Metto and Petra Telgkamp and Autumn K. McLane-Svoboda and Assaf A. Gilad and Galit Pelled}, url = {https://www.sciencedirect.com/science/article/pii/S0006899323003621}, doi = {https://doi.org/10.1016/j.brainres.2023.148591}, year = {2023}, date = {2023-09-24}, journal = {Brain Research}, abstract = {On-demand neurostimulation has shown success in epilepsy patients with pharmacoresistant seizures. Seizures produce magnetic fields that can be recorded using magnetoencephalography. We developed a new closed-loop approach to control seizure activity based on magnetogenetics using the electromagnetic perceptive gene (EPG) that encodes a protein that responds to magnetic fields. The EPG transgene was expressed in inhibitory interneurons under the hDlx promoter and kainic acid was used to induce acute seizures. In vivo electrophysiological signals were recorded. We found that hDlx EPG rats exhibited a significant delay in the onset of first seizure (1142.72 ± 186.35 s) compared to controls (644.03 ± 15.06 s) and significantly less seizures (4.11 ± 1.03) compared to controls (8.33 ± 1.58). These preliminary findings suggest that on-demand activation of EPG expressed in inhibitory interneurons suppresses seizure activity, and magnetogenetics via EPG may be an effective strategy to alleviate seizure severity in a closed-loop, and cell-specific fashion.}, keywords = {}, pubstate = {published}, tppubtype = {article} } On-demand neurostimulation has shown success in epilepsy patients with pharmacoresistant seizures. Seizures produce magnetic fields that can be recorded using magnetoencephalography. We developed a new closed-loop approach to control seizure activity based on magnetogenetics using the electromagnetic perceptive gene (EPG) that encodes a protein that responds to magnetic fields. The EPG transgene was expressed in inhibitory interneurons under the hDlx promoter and kainic acid was used to induce acute seizures. In vivo electrophysiological signals were recorded. We found that hDlx EPG rats exhibited a significant delay in the onset of first seizure (1142.72 ± 186.35 s) compared to controls (644.03 ± 15.06 s) and significantly less seizures (4.11 ± 1.03) compared to controls (8.33 ± 1.58). These preliminary findings suggest that on-demand activation of EPG expressed in inhibitory interneurons suppresses seizure activity, and magnetogenetics via EPG may be an effective strategy to alleviate seizure severity in a closed-loop, and cell-specific fashion. |
  | Levi, Timothee; Beaubois, Romain; Cheslet, Jérémy; Duenki, Tomoya; Khoyratee, Farad; Branchereau, Pascal; Ikeuchi, Yoshiho BiœmuS: A new tool for neurological disorders studies through real-time emulation and hybridization using biomimetic Spiking Neural Network Journal Article Research Square, 2023. @article{Levi2023, title = {BiœmuS: A new tool for neurological disorders studies through real-time emulation and hybridization using biomimetic Spiking Neural Network}, author = {Timothee Levi and Romain Beaubois and Jérémy Cheslet and Tomoya Duenki and Farad Khoyratee and Pascal Branchereau and Yoshiho Ikeuchi}, url = {https://www.researchsquare.com}, doi = {10.21203/rs.3.rs-3191285/v1}, year = {2023}, date = {2023-09-15}, journal = {Research Square}, abstract = {Characterization and modeling of biological neural networks is a field opening to major advances in our understanding of the mechanisms governing the functioning of the brain and the different pathologies that can affect it. Recent researches in bioelectronics and neuromorphic engineering lead to the design of the new generation of neuroprosthesis. Here we show a novel real-time, biomimetic and energy-efficient neural network for bio-hybrid experiments and parallel emulation. This novel system is used to investigate and reproduce neural network dynamics. The setup is running on a digital platform using a System on Chip (SoC) featuring both Programmable Logic (PL) and processors in a Processing System (PS) part. The FPGA part is computing the biomimetic and real-time electrical activities of Hodgkin-Huxley neural network while the processors handle monitoring and communication. New methods of resource and power optimization has been applied to the FPGA to allow detailed neuron modeling with synapses showing short term plasticity. The system is validated by comparison with biological data and model. We also demonstrate the feasibility of bio-hybrid experiments with different bio-physical interface and different biological cells. The complete setup achieves communication with a fully flexible real-time device thus constituting a step towards neuromorphic-based neuroprosthesis for bioelectrical therapeutics.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Characterization and modeling of biological neural networks is a field opening to major advances in our understanding of the mechanisms governing the functioning of the brain and the different pathologies that can affect it. Recent researches in bioelectronics and neuromorphic engineering lead to the design of the new generation of neuroprosthesis. Here we show a novel real-time, biomimetic and energy-efficient neural network for bio-hybrid experiments and parallel emulation. This novel system is used to investigate and reproduce neural network dynamics. The setup is running on a digital platform using a System on Chip (SoC) featuring both Programmable Logic (PL) and processors in a Processing System (PS) part. The FPGA part is computing the biomimetic and real-time electrical activities of Hodgkin-Huxley neural network while the processors handle monitoring and communication. New methods of resource and power optimization has been applied to the FPGA to allow detailed neuron modeling with synapses showing short term plasticity. The system is validated by comparison with biological data and model. We also demonstrate the feasibility of bio-hybrid experiments with different bio-physical interface and different biological cells. The complete setup achieves communication with a fully flexible real-time device thus constituting a step towards neuromorphic-based neuroprosthesis for bioelectrical therapeutics. |
  | Silverman, Jill L; Fenton, Timothy; Haouchine, Olivia; Hallam, Elizabeth; Smith, Emily; Jackson, Kiya; Rahbarian, Darlene; Canales, Cesar; Adhikari, Anna; Nord, Alex; Ben-Shalom, Roy Hyperexcitability and translational phenotypes in a preclinical model of SYNGAP1mutations Journal Article Research Square, 2023. @article{Silverman2023, title = {Hyperexcitability and translational phenotypes in a preclinical model of SYNGAP1mutations}, author = {Jill L. Silverman and Timothy Fenton and Olivia Haouchine and Elizabeth Hallam and Emily Smith and Kiya Jackson and Darlene Rahbarian and Cesar Canales and Anna Adhikari and Alex Nord and Roy Ben-Shalom}, url = {https://www.researchsquare.com/article/rs-3246655/v1}, doi = {https://doi.org/10.21203/rs.3.rs-3246655/v1}, year = {2023}, date = {2023-09-13}, journal = {Research Square}, abstract = {SYNGAP1is a critical gene for neuronal development, synaptic structure, and function. Although rare, the disruption of SYNGAP1directly causes a genetically identi able neurodevelopmental disorder (NDD) called SYNGAP1-related intellectual disability. Without functional SynGAP1 protein, patients present with intellectual disability, motor impairments, and epilepsy. Previous work using mouse models with a variety of germline and conditional mutations has helped delineate SynGAP1’s critical roles in neuronal structure and function, as well as key biochemical signaling pathways essential to synapse integrity. Homozygous loss of SYNGAP1is embryonically lethal. Heterozygous mutations of SynGAP1result in a broad range of phenotypes including increased locomotor activity, impaired working spatial memory, impaired cued fear memory, and increased stereotypic behavior. Ourinvivofunctional data, using the original germline mutation mouse line from the Huganir laboratory, corroborated robust hyperactivity and learning and memory de cits. Here, we describe impairments in the translational biomarker domain of sleep, characterized using neurophysiological data collected with wireless telemetric electroencephalography (EEG). We discoveredSyngap1+/− mice exhibited elevated spike trains in both number and duration, in addition to elevated power, most notably in the delta power band. Primary neurons fromSyngap1+/− mice displayed increased network ring activity, greater spikes per burst, and shorter inter-burst intervals between peaks using high density micro-electrode arrays (HD-MEA). This work is translational, innovative, and highly signi cant as it outlines functional impairments in Syngap1mutant mice. Simultaneously, the work utilized untethered, wireless neurophysiology that can discover potential biomarkers of Syngap1RID, for clinical trials, as it has done with other NDDs. Our work is substantial forward progress toward translational work for SynGAP1R-ID as it bridges in-vitroelectrophysiological neuronal activity and function with invivoneurophysiological brain activity and function. These data elucidate multiple quantitative, translational biomarkers invivoand invitrofor the development of treatments for SYNGAP1-related intellectual disability.}, keywords = {}, pubstate = {published}, tppubtype = {article} } SYNGAP1is a critical gene for neuronal development, synaptic structure, and function. Although rare, the disruption of SYNGAP1directly causes a genetically identi able neurodevelopmental disorder (NDD) called SYNGAP1-related intellectual disability. Without functional SynGAP1 protein, patients present with intellectual disability, motor impairments, and epilepsy. Previous work using mouse models with a variety of germline and conditional mutations has helped delineate SynGAP1’s critical roles in neuronal structure and function, as well as key biochemical signaling pathways essential to synapse integrity. Homozygous loss of SYNGAP1is embryonically lethal. Heterozygous mutations of SynGAP1result in a broad range of phenotypes including increased locomotor activity, impaired working spatial memory, impaired cued fear memory, and increased stereotypic behavior. Ourinvivofunctional data, using the original germline mutation mouse line from the Huganir laboratory, corroborated robust hyperactivity and learning and memory de cits. Here, we describe impairments in the translational biomarker domain of sleep, characterized using neurophysiological data collected with wireless telemetric electroencephalography (EEG). We discoveredSyngap1+/− mice exhibited elevated spike trains in both number and duration, in addition to elevated power, most notably in the delta power band. Primary neurons fromSyngap1+/− mice displayed increased network ring activity, greater spikes per burst, and shorter inter-burst intervals between peaks using high density micro-electrode arrays (HD-MEA). This work is translational, innovative, and highly signi cant as it outlines functional impairments in Syngap1mutant mice. Simultaneously, the work utilized untethered, wireless neurophysiology that can discover potential biomarkers of Syngap1RID, for clinical trials, as it has done with other NDDs. Our work is substantial forward progress toward translational work for SynGAP1R-ID as it bridges in-vitroelectrophysiological neuronal activity and function with invivoneurophysiological brain activity and function. These data elucidate multiple quantitative, translational biomarkers invivoand invitrofor the development of treatments for SYNGAP1-related intellectual disability. |
  | Habibollahi, Forough; Kagan, Brett J; Burkitt, Anthony N; French, Chris Critical dynamics arise during structured information presentation within embodied in vitro neuronal networks Journal Article Nature Communications, 2023. @article{Habibollahi2023, title = {Critical dynamics arise during structured information presentation within embodied in vitro neuronal networks}, author = {Forough Habibollahi and Brett J. Kagan and Anthony N. Burkitt and Chris French }, url = {https://www.nature.com/articles/s41467-023-41020-3}, doi = {https://doi.org/10.1038/s41467-023-41020-3}, year = {2023}, date = {2023-08-30}, journal = {Nature Communications}, abstract = {Understanding how brains process information is an incredibly difficult task. Amongst the metrics characterising information processing in the brain, observations of dynamic near-critical states have generated significant interest. However, theoretical and experimental limitations associated with human and animal models have precluded a definite answer about when and why neural criticality arises with links from attention, to cognition, and even to consciousness. To explore this topic, we used an in vitro neural network of cortical neurons that was trained to play a simplified game of ‘Pong’ to demonstrate Synthetic Biological Intelligence (SBI). We demonstrate that critical dynamics emerge when neural networks receive task-related structured sensory input, reorganizing the system to a near-critical state. Additionally, better task performance correlated with proximity to critical dynamics. However, criticality alone is insufficient for a neuronal network to demonstrate learning in the absence of additional information regarding the consequences of previous actions. These findings offer compelling support that neural criticality arises as a base feature of incoming structured information processing without the need for higher order cognition.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Understanding how brains process information is an incredibly difficult task. Amongst the metrics characterising information processing in the brain, observations of dynamic near-critical states have generated significant interest. However, theoretical and experimental limitations associated with human and animal models have precluded a definite answer about when and why neural criticality arises with links from attention, to cognition, and even to consciousness. To explore this topic, we used an in vitro neural network of cortical neurons that was trained to play a simplified game of ‘Pong’ to demonstrate Synthetic Biological Intelligence (SBI). We demonstrate that critical dynamics emerge when neural networks receive task-related structured sensory input, reorganizing the system to a near-critical state. Additionally, better task performance correlated with proximity to critical dynamics. However, criticality alone is insufficient for a neuronal network to demonstrate learning in the absence of additional information regarding the consequences of previous actions. These findings offer compelling support that neural criticality arises as a base feature of incoming structured information processing without the need for higher order cognition. |
  | Radivojevic, Milos; Punga, Anna Rostedt Functional imaging of conduction dynamics in cortical and spinal axons Journal Article eLife, 2023. @article{Radivojevic2023_2, title = {Functional imaging of conduction dynamics in cortical and spinal axons}, author = {Milos Radivojevic and Anna Rostedt Punga}, url = {https://elifesciences.org/articles/86512}, doi = {https://doi.org/10.7554/eLife.86512}, year = {2023}, date = {2023-08-22}, journal = {eLife}, abstract = {Mammalian axons are specialized for transmitting action potentials to targets within the central and peripheral nervous system. A growing body of evidence suggests that, besides signal conduction, axons play essential roles in neural information processing, and their malfunctions are common hallmarks of neurodegenerative diseases. The technologies available to study axonal function and structure integrally limit the comprehension of axon neurobiology. High-density microelectrode arrays (HD-MEAs) allow for accessing axonal action potentials at high spatiotemporal resolution, but provide no insights on axonal morphology. Here, we demonstrate a method for electrical visualization of axonal morphologies based on extracellular action potentials recorded from cortical and motor neurons using HD-MEAs. The method enabled us to reconstruct up to 5-cm-long axonal arbors and directly monitor axonal conduction across thousands of recording sites. We reconstructed 1.86 m of cortical and spinal axons in total and found specific features in their structure and function.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Mammalian axons are specialized for transmitting action potentials to targets within the central and peripheral nervous system. A growing body of evidence suggests that, besides signal conduction, axons play essential roles in neural information processing, and their malfunctions are common hallmarks of neurodegenerative diseases. The technologies available to study axonal function and structure integrally limit the comprehension of axon neurobiology. High-density microelectrode arrays (HD-MEAs) allow for accessing axonal action potentials at high spatiotemporal resolution, but provide no insights on axonal morphology. Here, we demonstrate a method for electrical visualization of axonal morphologies based on extracellular action potentials recorded from cortical and motor neurons using HD-MEAs. The method enabled us to reconstruct up to 5-cm-long axonal arbors and directly monitor axonal conduction across thousands of recording sites. We reconstructed 1.86 m of cortical and spinal axons in total and found specific features in their structure and function. |
  | Duru, Jens; Maurer, Benedikt; Doran, Ciara Giles; Jelitto, Robert; Küchler, Joël; Ihle, Stephan J; Ruff, Tobias; John, Robert; Genocchi, Barbara; Vörös, János Investigation of the input-output relationship of engineered neural networks using high-density microelectrode arrays Journal Article Biosensors and Bioelectronics, 2023. @article{Duru2023b, title = {Investigation of the input-output relationship of engineered neural networks using high-density microelectrode arrays}, author = {Jens Duru and Benedikt Maurer and Ciara Giles Doran and Robert Jelitto and Joël Küchler and Stephan J. Ihle and Tobias Ruff and Robert John and Barbara Genocchi and János Vörös }, url = {https://www.sciencedirect.com/science/article/pii/S095656632300533X?via%3Dihub}, doi = {https://doi.org/10.1016/j.bios.2023.115591}, year = {2023}, date = {2023-08-18}, journal = {Biosensors and Bioelectronics}, abstract = {Bottom-up neuroscience utilizes small, engineered biological neural networks to study neuronal activity in systems of reduced complexity. We present a platform that establishes up to six independent networks formed by primary rat neurons on planar complementary metal–oxide–semiconductor (CMOS) microelectrode arrays (MEAs). We introduce an approach that allows repetitive stimulation and recording of network activity at any of the over 700 electrodes underlying a network. We demonstrate that the continuous application of a repetitive super-threshold stimulus yields a reproducible network answer within a 15 ms post-stimulus window. This response can be tracked with high spatiotemporal resolution across the whole extent of the network. Moreover, we show that the location of the stimulation plays a significant role in the networks' early response to the stimulus. By applying a stimulation pattern to all network-underlying electrodes in sequence, the sensitivity of the whole network to the stimulus can be visualized. We demonstrate that microchannels reduce the voltage stimulation threshold and induce the strongest network response. By varying the stimulation amplitude and frequency we reveal discrete network transition points. Finally, we introduce vector fields to follow stimulation-induced spike propagation pathways within the network. Overall we show that our defined neural networks on CMOS MEAs enable us to elicit highly reproducible activity patterns that can be precisely modulated by stimulation amplitude, stimulation frequency and the site of stimulation.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Bottom-up neuroscience utilizes small, engineered biological neural networks to study neuronal activity in systems of reduced complexity. We present a platform that establishes up to six independent networks formed by primary rat neurons on planar complementary metal–oxide–semiconductor (CMOS) microelectrode arrays (MEAs). We introduce an approach that allows repetitive stimulation and recording of network activity at any of the over 700 electrodes underlying a network. We demonstrate that the continuous application of a repetitive super-threshold stimulus yields a reproducible network answer within a 15 ms post-stimulus window. This response can be tracked with high spatiotemporal resolution across the whole extent of the network. Moreover, we show that the location of the stimulation plays a significant role in the networks' early response to the stimulus. By applying a stimulation pattern to all network-underlying electrodes in sequence, the sensitivity of the whole network to the stimulus can be visualized. We demonstrate that microchannels reduce the voltage stimulation threshold and induce the strongest network response. By varying the stimulation amplitude and frequency we reveal discrete network transition points. Finally, we introduce vector fields to follow stimulation-induced spike propagation pathways within the network. Overall we show that our defined neural networks on CMOS MEAs enable us to elicit highly reproducible activity patterns that can be precisely modulated by stimulation amplitude, stimulation frequency and the site of stimulation. |
  | Yamamoto, Hideaki; Hirano-Iwata, Ayumi; Sato, Shigeo Microfluidic technologies for reconstituting neuronal network functions in vitro Journal Article JSAP Review, 2023. @article{Yamamoto2023, title = {Microfluidic technologies for reconstituting neuronal network functions in vitro}, author = {Hideaki Yamamoto and Ayumi Hirano-Iwata and Shigeo Sato}, url = {https://www.jstage.jst.go.jp/article/oubutsu/92/5/92_278/_article/-char/en}, doi = {10.11470/oubutsu.92.5_278}, year = {2023}, date = {2023-07-06}, journal = {JSAP Review}, abstract = {The structure and function of complex neuronal networks in the brain can be partially reconstituted in vitro by integrating cell culture and microfluidic device technologies. In this report, we review our recent studies on developing microfluidic devices to reconstitute small neuronal networks bearing a modular structure, which is a canonical structure found in the nervous systems of animals. We also describe the process of recording functional activity from the reconstituted neuronal networks. These fundamental technologies offer novel tools for investigating structure–function relationships in living neuronal networks and exploring the physical basis of biological computing in the brain.}, keywords = {}, pubstate = {published}, tppubtype = {article} } The structure and function of complex neuronal networks in the brain can be partially reconstituted in vitro by integrating cell culture and microfluidic device technologies. In this report, we review our recent studies on developing microfluidic devices to reconstitute small neuronal networks bearing a modular structure, which is a canonical structure found in the nervous systems of animals. We also describe the process of recording functional activity from the reconstituted neuronal networks. These fundamental technologies offer novel tools for investigating structure–function relationships in living neuronal networks and exploring the physical basis of biological computing in the brain. |
  | Zhao, Eric T; Hull, Jacob M; Hemed, Nofar Mintz; Ulusan, Hasan; Bartram, Julian; Zhang, Anqi; Wang, Pingyu; Pham, Albert; Silvia Ronchi, John Huguenard R; Hierlemann, Andreas; Melosh, Nicholas A A CMOS-based highly scalable flexible neural electrode interface Journal Article Science Advances, 2023. @article{Zhao2023, title = {A CMOS-based highly scalable flexible neural electrode interface}, author = {Eric T. Zhao and Jacob M. Hull and Nofar Mintz Hemed and Hasan Ulusan and Julian Bartram and Anqi Zhang and Pingyu Wang and Albert Pham and Silvia Ronchi, John R. Huguenard and Andreas Hierlemann and Nicholas A. Melosh}, url = {https://www.science.org/doi/10.1126/sciadv.adf9524}, doi = {DOI: 10.1126/sciadv.adf9524}, year = {2023}, date = {2023-06-07}, journal = {Science Advances}, abstract = {Perception, thoughts, and actions are encoded by the coordinated activity of large neuronal populations spread over large areas. However, existing electrophysiological devices are limited by their scalability in capturing this cortex-wide activity. Here, we developed an electrode connector based on an ultra-conformable thin-film electrode array that self-assembles onto silicon microelectrode arrays enabling multithousand channel counts at a millimeter scale. The interconnects are formed using microfabricated electrode pads suspended by thin support arms, termed Flex2Chip. Capillary-assisted assembly drives the pads to deform toward the chip surface, and van der Waals forces maintain this deformation, establishing Ohmic contact. Flex2Chip arrays successfully measured extracellular action potentials ex vivo and resolved micrometer scale seizure propagation trajectories in epileptic mice. We find that seizure dynamics in absence epilepsy in the Scn8a+/− model do not have constant propagation trajectories.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Perception, thoughts, and actions are encoded by the coordinated activity of large neuronal populations spread over large areas. However, existing electrophysiological devices are limited by their scalability in capturing this cortex-wide activity. Here, we developed an electrode connector based on an ultra-conformable thin-film electrode array that self-assembles onto silicon microelectrode arrays enabling multithousand channel counts at a millimeter scale. The interconnects are formed using microfabricated electrode pads suspended by thin support arms, termed Flex2Chip. Capillary-assisted assembly drives the pads to deform toward the chip surface, and van der Waals forces maintain this deformation, establishing Ohmic contact. Flex2Chip arrays successfully measured extracellular action potentials ex vivo and resolved micrometer scale seizure propagation trajectories in epileptic mice. We find that seizure dynamics in absence epilepsy in the Scn8a+/− model do not have constant propagation trajectories. |
  | Girardi, Gregory; Zumpano, Danielle; Goshi, Noah; Raybould, Helen; Seker, Erkin Cultured Vagal Afferent Neurons as Sensors for Intestinal Effector Molecules Journal Article biosensors, 2023. @article{Girardi2023, title = {Cultured Vagal Afferent Neurons as Sensors for Intestinal Effector Molecules}, author = {Gregory Girardi and Danielle Zumpano and Noah Goshi and Helen Raybould and Erkin Seker}, url = {https://www.mdpi.com/2079-6374/13/6/601}, doi = {10.3390/bios13060601}, year = {2023}, date = {2023-05-31}, journal = {biosensors}, abstract = {The gut–brain axis embodies the bi-directional communication between the gastrointestinal tract and the central nervous system (CNS), where vagal afferent neurons (VANs) serve as sensors for a variety of gut-derived signals. The gut is colonized by a large and diverse population of microorganisms that communicate via small (effector) molecules, which also act on the VAN terminals situated in the gut viscera and consequently influence many CNS processes. However, the convoluted in vivo environment makes it difficult to study the causative impact of the effector molecules on VAN activation or desensitization. Here, we report on a VAN culture and its proof-of-principle demonstration as a cell-based sensor to monitor the influence of gastrointestinal effector molecules on neuronal behavior. We initially compared the effect of surface coatings (poly-L-lysine vs. Matrigel) and culture media composition (serum vs. growth factor supplement) on neurite growth as a surrogate of VAN regeneration following tissue harvesting, where the Matrigel coating, but not the media composition, played a significant role in the increased neurite growth. We then used both live-cell calcium imaging and extracellular electrophysiological recordings to show that the VANs responded to classical effector molecules of endogenous and exogenous origin (cholecystokinin serotonin and capsaicin) in a complex fashion. We expect this study to enable platforms for screening various effector molecules and their influence on VAN activity, assessed by their information-rich electrophysiological fingerprints.}, keywords = {}, pubstate = {published}, tppubtype = {article} } The gut–brain axis embodies the bi-directional communication between the gastrointestinal tract and the central nervous system (CNS), where vagal afferent neurons (VANs) serve as sensors for a variety of gut-derived signals. The gut is colonized by a large and diverse population of microorganisms that communicate via small (effector) molecules, which also act on the VAN terminals situated in the gut viscera and consequently influence many CNS processes. However, the convoluted in vivo environment makes it difficult to study the causative impact of the effector molecules on VAN activation or desensitization. Here, we report on a VAN culture and its proof-of-principle demonstration as a cell-based sensor to monitor the influence of gastrointestinal effector molecules on neuronal behavior. We initially compared the effect of surface coatings (poly-L-lysine vs. Matrigel) and culture media composition (serum vs. growth factor supplement) on neurite growth as a surrogate of VAN regeneration following tissue harvesting, where the Matrigel coating, but not the media composition, played a significant role in the increased neurite growth. We then used both live-cell calcium imaging and extracellular electrophysiological recordings to show that the VANs responded to classical effector molecules of endogenous and exogenous origin (cholecystokinin serotonin and capsaicin) in a complex fashion. We expect this study to enable platforms for screening various effector molecules and their influence on VAN activity, assessed by their information-rich electrophysiological fingerprints. |
  | Bartram, Julian; Franke, Felix; Kumar, Sreedhar Saseendran; Buccino, Alessio Paolo; Xue, Xiaohan; Gänswein, Tobias; Schröter, Manuel; Kim, Taehoon; Kasuba, Krishna Chaitanya; Hierlemann, Andreas Parallel reconstruction of the excitatory and inhibitory inputs received by single neurons reveals the synaptic basis of recurrent spiking Journal Article eLife, 2023. @article{Bartram2023b, title = {Parallel reconstruction of the excitatory and inhibitory inputs received by single neurons reveals the synaptic basis of recurrent spiking}, author = {Julian Bartram and Felix Franke and Sreedhar Saseendran Kumar and Alessio Paolo Buccino and Xiaohan Xue and Tobias Gänswein and Manuel Schröter and Taehoon Kim and Krishna Chaitanya Kasuba and Andreas Hierlemann}, url = {https://elifesciences.org/reviewed-preprints/86820}, doi = {10.7554/eLife.86820}, year = {2023}, date = {2023-05-17}, journal = {eLife}, abstract = {Self-sustained recurrent activity in cortical networks is thought to be important for multiple crucial processes, including circuit development and homeostasis. Yet, the precise relationship between the synaptic input patterns and the spiking output of individual neurons remains largely unresolved. Here, we developed, validated and applied a novel in vitro experimental platform and analytical procedures that provide – for individual neurons – simultaneous excitatory and inhibitory synaptic activity estimates during recurrent network activity. Our approach combines whole-network high-density microelectrode array (HD-MEA) recordings from rat neuronal cultures with patch clamping and enables a comprehensive mapping and characterization of active incoming connections to single postsynaptic neurons. We found that, during network states with excitation(E)-inhibition(I) balance, postsynaptic spiking coincided precisely with the maxima of fast fluctuations in the input E/I ratio. These spike-associated E/I ratio escalations were largely due to a rapid bidirectional change in synaptic inhibition that was modulated by the network-activity level. Our approach also uncovered the underlying circuit architecture and we show that individual neurons received a few key inhibitory connections – often from special hub neurons – that were instrumental in controlling postsynaptic spiking. Balanced network theory predicts dynamical regimes governed by small and rapid input fluctuation and featuring a fast neuronal responsiveness. Our findings – obtained in self-organized neuronal cultures – suggest that the emergence of these favorable regimes and associated network architectures is an inherent property of cortical networks in general.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Self-sustained recurrent activity in cortical networks is thought to be important for multiple crucial processes, including circuit development and homeostasis. Yet, the precise relationship between the synaptic input patterns and the spiking output of individual neurons remains largely unresolved. Here, we developed, validated and applied a novel in vitro experimental platform and analytical procedures that provide – for individual neurons – simultaneous excitatory and inhibitory synaptic activity estimates during recurrent network activity. Our approach combines whole-network high-density microelectrode array (HD-MEA) recordings from rat neuronal cultures with patch clamping and enables a comprehensive mapping and characterization of active incoming connections to single postsynaptic neurons. We found that, during network states with excitation(E)-inhibition(I) balance, postsynaptic spiking coincided precisely with the maxima of fast fluctuations in the input E/I ratio. These spike-associated E/I ratio escalations were largely due to a rapid bidirectional change in synaptic inhibition that was modulated by the network-activity level. Our approach also uncovered the underlying circuit architecture and we show that individual neurons received a few key inhibitory connections – often from special hub neurons – that were instrumental in controlling postsynaptic spiking. Balanced network theory predicts dynamical regimes governed by small and rapid input fluctuation and featuring a fast neuronal responsiveness. Our findings – obtained in self-organized neuronal cultures – suggest that the emergence of these favorable regimes and associated network architectures is an inherent property of cortical networks in general. |
  | Duru, Jens; Maurer, Benedikt; Doran, Ciara Giles; Jelitto, Robert; Küchler, Joël; Ihle, Stephan J; Ruff, Tobias; John, Robert; Genocchi, Barbara; Vörös, János Investigation of the input-output relationship of engineered neural networks using high-density microelectrode arrays Journal Article SSRN, 2023. @article{Duru2023, title = {Investigation of the input-output relationship of engineered neural networks using high-density microelectrode arrays}, author = {Jens Duru and Benedikt Maurer and Ciara Giles Doran and Robert Jelitto and Joël Küchler and Stephan J. Ihle and Tobias Ruff and Robert John and Barbara Genocchi and János Vörös}, url = {https://www.ssrn.com/abstract=4427959}, doi = {DOI: 10.2139/ssrn.4427959}, year = {2023}, date = {2023-04-24}, journal = {SSRN}, abstract = {Bottom-up neuroscience utilizes small, engineered biological neural networks to study neuronal activity in systems of reduced complexity. We present a platform that establishes up to six independent networks formed by primary rat neurons on planar complementary metal–oxide–semiconductor (CMOS) microelectrode arrays (MEAs). We introduce an approach that allows repetitive stimulation and recording of network activity at any of the over 700 electrodes underlying a network. We demonstrate that the continuous application of a repetitive super-threshold stimulus yields a reproducible network answer within a 15 ms post-stimulus window. This response can be tracked with high spatiotemporal resolution across the whole extent of the network. Moreover, we show that the location of the stimulation plays a significant role in the networks’ early response to the stimulus. By applying a stimulation pattern to all network-underlying electrodes in sequence, the sensitivity of the whole network to the stimulus can be visualized. We demonstrate that microchannels reduce the voltage stimulation threshold and induce the strongest network response. By varying the stimulation amplitude and frequency we reveal discrete network transition points. Finally, we introduce vector fields to follow stimulation-induced spike propagation pathways within the network. Overall we show that our defined neural networks on CMOS MEAs enable us to elicit highly reproducible activity patterns that can be precisely modulated by stimulation amplitude, stimulation frequency and the site of stimulation.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Bottom-up neuroscience utilizes small, engineered biological neural networks to study neuronal activity in systems of reduced complexity. We present a platform that establishes up to six independent networks formed by primary rat neurons on planar complementary metal–oxide–semiconductor (CMOS) microelectrode arrays (MEAs). We introduce an approach that allows repetitive stimulation and recording of network activity at any of the over 700 electrodes underlying a network. We demonstrate that the continuous application of a repetitive super-threshold stimulus yields a reproducible network answer within a 15 ms post-stimulus window. This response can be tracked with high spatiotemporal resolution across the whole extent of the network. Moreover, we show that the location of the stimulation plays a significant role in the networks’ early response to the stimulus. By applying a stimulation pattern to all network-underlying electrodes in sequence, the sensitivity of the whole network to the stimulus can be visualized. We demonstrate that microchannels reduce the voltage stimulation threshold and induce the strongest network response. By varying the stimulation amplitude and frequency we reveal discrete network transition points. Finally, we introduce vector fields to follow stimulation-induced spike propagation pathways within the network. Overall we show that our defined neural networks on CMOS MEAs enable us to elicit highly reproducible activity patterns that can be precisely modulated by stimulation amplitude, stimulation frequency and the site of stimulation. |
  | Xu, He Jax; Yao, Yao; Yao, Fenyong; Chen, Jiehui; Li, Meishi; Yang, Xianfa; Li, Sheng; Lu, Fangru; Hu, Ping; He, Shuijin; Peng, Guangdun; Jing, Naihe Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells Journal Article Cell Regeneration, 2023. @article{Xu2023, title = {Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells}, author = {He Jax Xu and Yao Yao and Fenyong Yao and Jiehui Chen and Meishi Li and Xianfa Yang and Sheng Li and Fangru Lu and Ping Hu and Shuijin He and Guangdun Peng and Naihe Jing}, url = {https://cellregeneration.springeropen.com/articles/10.1186/s13619-023-00159-6}, doi = {10.1186/s13619-023-00159-6}, year = {2023}, date = {2023-03-23}, journal = {Cell Regeneration}, abstract = {Spinal motor neurons deficiency results in a series of devastating disorders such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA) and spinal cord injury (SCI). These disorders are currently incurable, while human pluripotent stem cells (hPSCs)-derived spinal motor neurons are promising but suffered from inappropriate regional identity and functional immaturity for the study and treatment of posterior spinal cord related injuries. In this study, we have established human spinal cord neural progenitor cells (hSCNPCs) via hPSCs differentiated neuromesodermal progenitors (NMPs) and demonstrated the hSCNPCs can be continuously expanded up to 40 passages. hSCNPCs can be rapidly differentiated into posterior spinal motor neurons with high efficiency. The functional maturity has been examined in detail. Moreover, a co-culture scheme which is compatible for both neural and muscular differentiation is developed to mimic the neuromuscular junction (NMJ) formation in vitro. Together, these studies highlight the potential avenues for generating clinically relevant spinal motor neurons and modeling neuromuscular diseases through our defined hSCNPCs.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Spinal motor neurons deficiency results in a series of devastating disorders such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA) and spinal cord injury (SCI). These disorders are currently incurable, while human pluripotent stem cells (hPSCs)-derived spinal motor neurons are promising but suffered from inappropriate regional identity and functional immaturity for the study and treatment of posterior spinal cord related injuries. In this study, we have established human spinal cord neural progenitor cells (hSCNPCs) via hPSCs differentiated neuromesodermal progenitors (NMPs) and demonstrated the hSCNPCs can be continuously expanded up to 40 passages. hSCNPCs can be rapidly differentiated into posterior spinal motor neurons with high efficiency. The functional maturity has been examined in detail. Moreover, a co-culture scheme which is compatible for both neural and muscular differentiation is developed to mimic the neuromuscular junction (NMJ) formation in vitro. Together, these studies highlight the potential avenues for generating clinically relevant spinal motor neurons and modeling neuromuscular diseases through our defined hSCNPCs. |
  | Radivojevic, Milos; Punga, Anna Rostedt Functional imaging of conduction dynamics in cortical and spinal axons Journal Article BioRxiv, 2023. @article{Radivojevic2023, title = {Functional imaging of conduction dynamics in cortical and spinal axons}, author = {Milos Radivojevic and Anna Rostedt Punga}, url = {https://www.biorxiv.org/content/10.1101/2023.02.28.530461v1}, doi = {https://doi.org/10.1101/2023.02.28.530461}, year = {2023}, date = {2023-03-01}, journal = {BioRxiv}, abstract = {Mammalian axons are specialized for transmitting action potentials to targets within the central and peripheral nervous system. A growing body of evidence suggests that, besides signal conduction, axons play essential roles in neural information processing, and their malfunctions are common hallmarks of neurodegenerative diseases. The technologies available to study axonal function and structure integrally limit the comprehension of axon neurobiology. High-density microelectrode arrays (HD-MEAs) allow for accessing axonal action potentials at high spatiotemporal resolution, but provide no insights on axonal morphology. Here we demonstrate a method for electrical visualization of axonal morphologies based on extracellular action potentials recorded from cortical and motor neurons using HD-MEAs. The method enabled us to reconstruct up to 5-centimeter-long axonal arbors and directly monitor axonal conduction across thousands of recording sites. We reconstructed 1.86 meters of cortical and spinal axons in total and found specific features in their structure and function.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Mammalian axons are specialized for transmitting action potentials to targets within the central and peripheral nervous system. A growing body of evidence suggests that, besides signal conduction, axons play essential roles in neural information processing, and their malfunctions are common hallmarks of neurodegenerative diseases. The technologies available to study axonal function and structure integrally limit the comprehension of axon neurobiology. High-density microelectrode arrays (HD-MEAs) allow for accessing axonal action potentials at high spatiotemporal resolution, but provide no insights on axonal morphology. Here we demonstrate a method for electrical visualization of axonal morphologies based on extracellular action potentials recorded from cortical and motor neurons using HD-MEAs. The method enabled us to reconstruct up to 5-centimeter-long axonal arbors and directly monitor axonal conduction across thousands of recording sites. We reconstructed 1.86 meters of cortical and spinal axons in total and found specific features in their structure and function. |
  | Cai, Hongwei; Ao, Zheng; Tian, Chunhui; Wu, Zhuhao; Liu, Hongcheng; Tchieu, Jason; Gu, Mingxia; Mackie, Ken; and Guo, Feng Brain Organoid Computing for Artificial Intelligence Journal Article bioRxiv, 2023. @article{Cai2023, title = {Brain Organoid Computing for Artificial Intelligence}, author = {Hongwei Cai and Zheng Ao and Chunhui Tian and Zhuhao Wu and Hongcheng Liu and Jason Tchieu and Mingxia Gu and Ken Mackie and and Feng Guo}, url = {https://www.biorxiv.org/content/10.1101/2023.02.28.530502v1}, doi = {10.1101/2023.02.28.530502}, year = {2023}, date = {2023-03-01}, journal = {bioRxiv}, abstract = {Brain-inspired hardware emulates the structure and working principles of a biological brain and may address the hardware bottleneck for fast-growing artificial intelligence (AI). Current brain-inspired silicon chips are promising but still limit their power to fully mimic brain function for AI computing. Here, we develop Brainoware, living AI hardware that harnesses the computation power of 3D biological neural networks in a brain organoid. Brain-like 3D in vitro cultures compute by receiving and sending information via a multielectrode array. Applying spatiotemporal electrical stimulation, this approach not only exhibits nonlinear dynamics and fading memory properties but also learns from training data. Further experiments demonstrate real-world applications in solving non-linear equations. This approach may provide new insights into AI hardware. }, keywords = {}, pubstate = {published}, tppubtype = {article} } Brain-inspired hardware emulates the structure and working principles of a biological brain and may address the hardware bottleneck for fast-growing artificial intelligence (AI). Current brain-inspired silicon chips are promising but still limit their power to fully mimic brain function for AI computing. Here, we develop Brainoware, living AI hardware that harnesses the computation power of 3D biological neural networks in a brain organoid. Brain-like 3D in vitro cultures compute by receiving and sending information via a multielectrode array. Applying spatiotemporal electrical stimulation, this approach not only exhibits nonlinear dynamics and fading memory properties but also learns from training data. Further experiments demonstrate real-world applications in solving non-linear equations. This approach may provide new insights into AI hardware. |
  | Kim, Eunhee; Jeon, Sungwoong; Yang, Yoon-Sil; Jin, Chaewon; Kim, Jin-young; Oh, Yong- Seok; Rah, Jong-Cheol; and Choi, Hongsoo A Neurospheroid-Based Microrobot for Targeted Neural Connections in a Hippocampal Slice Journal Article Advanced Materials, 2023. @article{EunheeKim2023, title = {A Neurospheroid-Based Microrobot for Targeted Neural Connections in a Hippocampal Slice}, author = {Eunhee Kim and Sungwoong Jeon and Yoon-Sil Yang and Chaewon Jin and Jin-young Kim and Yong- Seok Oh and Jong-Cheol Rah and and Hongsoo Choi}, url = {https://onlinelibrary.wiley.com/doi/10.1002/adma.202208747?af=R}, doi = {https://doi.org/10.1002/adma.202208747}, year = {2023}, date = {2023-01-14}, journal = {Advanced Materials}, abstract = {Functional restoration by the re-establishment of cellular or neural connections remains a major challenge in targeted cell therapy and regenerative medicine. Recent advances in magnetically powered microrobots have shown potential for use in controlled and targeted cell therapy. In this study, a magnetic neurospheroid (Mag-Neurobot) that could form both structural and functional connections with an organotypic hippocampal slice (OHS) was assessed using an ex vivo model as a bridge toward in vivo application. The Mag-Neurobot consists of hippocampal neurons and superparamagnetic nanoparticles (SPIONs); it is precisely and skillfully manipulated by an external magnetic field. Furthermore, the results of patch-clamp recordings of hippocampal neurons indicated that neither the neuronal excitabilities nor the synaptic functions of SPION-loaded cells were significantly affected. Analysis of neural activity propagation using high-density multi-electrode arrays showed that the delivered Mag-Neurobot was functionally connected with the OHS. The applications of this study include functional verification for targeted cell delivery through the characterization of novel synaptic connections and the functionalities of transported and transplanted cells. The success of the Mag-Neurobot opens up new avenues of research and application; it offers a test platform for functional neural connections and neural regenerative processes through cell transplantation.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Functional restoration by the re-establishment of cellular or neural connections remains a major challenge in targeted cell therapy and regenerative medicine. Recent advances in magnetically powered microrobots have shown potential for use in controlled and targeted cell therapy. In this study, a magnetic neurospheroid (Mag-Neurobot) that could form both structural and functional connections with an organotypic hippocampal slice (OHS) was assessed using an ex vivo model as a bridge toward in vivo application. The Mag-Neurobot consists of hippocampal neurons and superparamagnetic nanoparticles (SPIONs); it is precisely and skillfully manipulated by an external magnetic field. Furthermore, the results of patch-clamp recordings of hippocampal neurons indicated that neither the neuronal excitabilities nor the synaptic functions of SPION-loaded cells were significantly affected. Analysis of neural activity propagation using high-density multi-electrode arrays showed that the delivered Mag-Neurobot was functionally connected with the OHS. The applications of this study include functional verification for targeted cell delivery through the characterization of novel synaptic connections and the functionalities of transported and transplanted cells. The success of the Mag-Neurobot opens up new avenues of research and application; it offers a test platform for functional neural connections and neural regenerative processes through cell transplantation. |
  | Sato, Yuya; Yamamoto, Hideaki; Kato, Hideyuki; Tanii, Takashi; Sato, Shigeo; Hirano-Iwata, Ayumi Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks Journal Article Frontiers in Neuroscience, 2023. @article{Sato2023, title = {Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks}, author = {Yuya Sato and Hideaki Yamamoto and Hideyuki Kato and Takashi Tanii and Shigeo Sato and Ayumi Hirano-Iwata}, url = {https://www.frontiersin.org/articles/10.3389/fnins.2022.943310/full}, doi = {doi: 10.3389/fnins.2022.943310}, year = {2023}, date = {2023-01-09}, journal = {Frontiers in Neuroscience}, abstract = {Neuronal networks in dissociated culture combined with cell engineering technology offer a pivotal platform to constructively explore the relationship between structure and function in living neuronal networks. Here, we fabricated defined neuronal networks possessing a modular architecture on high-density microelectrode arrays (HD-MEAs), a state-of-the-art electrophysiological tool for recording neural activity with high spatial and temporal resolutions. We first established a surface coating protocol using a cell-permissive hydrogel to stably attach a polydimethylsiloxane microfluidic film on the HD-MEA. We then recorded the spontaneous neural activity of the engineered neuronal network, which revealed an important portrait of the engineered neuronal network–modular architecture enhances functional complexity by reducing the excessive neural correlation between spatially segregated modules. The results of this study highlight the impact of HD- MEA recordings combined with cell engineering technologies as a novel tool in neuroscience to constructively assess the structure-function relationships in neuronal networks.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Neuronal networks in dissociated culture combined with cell engineering technology offer a pivotal platform to constructively explore the relationship between structure and function in living neuronal networks. Here, we fabricated defined neuronal networks possessing a modular architecture on high-density microelectrode arrays (HD-MEAs), a state-of-the-art electrophysiological tool for recording neural activity with high spatial and temporal resolutions. We first established a surface coating protocol using a cell-permissive hydrogel to stably attach a polydimethylsiloxane microfluidic film on the HD-MEA. We then recorded the spontaneous neural activity of the engineered neuronal network, which revealed an important portrait of the engineered neuronal network–modular architecture enhances functional complexity by reducing the excessive neural correlation between spatially segregated modules. The results of this study highlight the impact of HD- MEA recordings combined with cell engineering technologies as a novel tool in neuroscience to constructively assess the structure-function relationships in neuronal networks. |
  | Han, Xiaobo; Matsuda, Naoki; Ishibashi, Yuto; Odawara, Aoi; Takahashi, Sayuri; Tooi, Norie; Kinoshita, Koshi; Suzuki, Ikuro A functional neuron maturation device provides convenient application on microelectrode array for neural network measurement Journal Article Biomaterials Research, 2022. @article{Han2022, title = {A functional neuron maturation device provides convenient application on microelectrode array for neural network measurement}, author = {Xiaobo Han and Naoki Matsuda and Yuto Ishibashi and Aoi Odawara and Sayuri Takahashi and Norie Tooi and Koshi Kinoshita and Ikuro Suzuki }, url = {https://biomaterialsres.biomedcentral.com/articles/10.1186/s40824-022-00324-z}, doi = {https://doi.org/10.1186/s40824-022-00324-z}, year = {2022}, date = {2022-12-20}, journal = {Biomaterials Research}, abstract = {Background Microelectrode array (MEA) systems are valuable for in vitro assessment of neurotoxicity and drug efficiency. However, several difficulties such as protracted functional maturation and high experimental costs hinder the use of MEA analysis requiring human induced pluripotent stem cells (hiPSCs). Neural network functional parameters are also needed for in vitro to in vivo extrapolation. Methods In the present study, we produced a cost effective nanofiber culture platform, the SCAD device, for long-term culture of hiPSC-derived neurons and primary peripheral neurons. The notable advantage of SCAD device is convenient application on multiple MEA systems for neuron functional analysis. Results We showed that the SCAD device could promote functional maturation of cultured hiPSC-derived neurons, and neurons responded appropriately to convulsant agents. Furthermore, we successfully analyzed parameters for in vitro to in vivo extrapolation, i.e., low-frequency components and synaptic propagation velocity of the signal, potentially reflecting neural network functions from neurons cultured on SCAD device. Finally, we measured the axonal conduction velocity of peripheral neurons. Conclusions: Neurons cultured on SCAD devices might constitute a reliable in vitro platform to investigate neuron functions, drug efficacy and toxicity, and neuropathological mechanisms by MEA.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Background Microelectrode array (MEA) systems are valuable for in vitro assessment of neurotoxicity and drug efficiency. However, several difficulties such as protracted functional maturation and high experimental costs hinder the use of MEA analysis requiring human induced pluripotent stem cells (hiPSCs). Neural network functional parameters are also needed for in vitro to in vivo extrapolation. Methods In the present study, we produced a cost effective nanofiber culture platform, the SCAD device, for long-term culture of hiPSC-derived neurons and primary peripheral neurons. The notable advantage of SCAD device is convenient application on multiple MEA systems for neuron functional analysis. Results We showed that the SCAD device could promote functional maturation of cultured hiPSC-derived neurons, and neurons responded appropriately to convulsant agents. Furthermore, we successfully analyzed parameters for in vitro to in vivo extrapolation, i.e., low-frequency components and synaptic propagation velocity of the signal, potentially reflecting neural network functions from neurons cultured on SCAD device. Finally, we measured the axonal conduction velocity of peripheral neurons. Conclusions: Neurons cultured on SCAD devices might constitute a reliable in vitro platform to investigate neuron functions, drug efficacy and toxicity, and neuropathological mechanisms by MEA. |
  | Tran, Hoang-Dai; Shin, Min-Kyoung; Denman, Charlotte; Han, Run-Run; Kuhn, Bernd; Arbuthnott, Gordon; Jo, Junghyun Generation of Human Striatal-Midbrain Assembloids From Human Pluripotent Stem Cells to Model Alpha-Synuclein Propagation Journal Article Sneak Peek - Cell Press, 2022. @article{Tran2022, title = {Generation of Human Striatal-Midbrain Assembloids From Human Pluripotent Stem Cells to Model Alpha-Synuclein Propagation}, author = {Hoang-Dai Tran and Min-Kyoung Shin and Charlotte Denman and Run-Run Han and Bernd Kuhn and Gordon Arbuthnott and Junghyun Jo}, url = {https://papers.ssrn.com/sol3/papers.cfm?abstract_id=4288935}, doi = {http://dx.doi.org/10.2139/ssrn.4288935}, year = {2022}, date = {2022-12-05}, journal = {Sneak Peek - Cell Press}, abstract = {Animal models of the pathology of Parkinson’s disease (PD) have provided most of the treatments to date, but the disease is restricted to human patients. In vitro models using human pluripotent stem cell-derived neural organoids have provided improved access to study PD etiology. Here, we generated human striatal and midbrain organoids and assembled both regionalized neural organoids to form human striatal-midbrain assembloids (hSMAs), mimicking a part of basal ganglia. Both the nigrostriatal and striatonigral pathways are present and electrophysiologically active in the hSMAs. hSMA development in the presence of increased alpha-synuclein (α-syn) from SNCA overexpression, induced nigrostriatal system damage, which is typical of the disease. Using the α-syn-mKO2 reporter and bimolecular fluorescence complementation system, we demonstrated that fluorescent α-syn is transported from the striatal area tothe dopaminergic (DA) neurons of the midbrain area. Furthermore, insoluble α-syn aggregated over time in DA neurons similar to Lewy bodies in human patients. Such assembloids are a compelling new platform to develop novel PD therapeutic strategies.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Animal models of the pathology of Parkinson’s disease (PD) have provided most of the treatments to date, but the disease is restricted to human patients. In vitro models using human pluripotent stem cell-derived neural organoids have provided improved access to study PD etiology. Here, we generated human striatal and midbrain organoids and assembled both regionalized neural organoids to form human striatal-midbrain assembloids (hSMAs), mimicking a part of basal ganglia. Both the nigrostriatal and striatonigral pathways are present and electrophysiologically active in the hSMAs. hSMA development in the presence of increased alpha-synuclein (α-syn) from SNCA overexpression, induced nigrostriatal system damage, which is typical of the disease. Using the α-syn-mKO2 reporter and bimolecular fluorescence complementation system, we demonstrated that fluorescent α-syn is transported from the striatal area tothe dopaminergic (DA) neurons of the midbrain area. Furthermore, insoluble α-syn aggregated over time in DA neurons similar to Lewy bodies in human patients. Such assembloids are a compelling new platform to develop novel PD therapeutic strategies. |
  | Akarca, Danyal; Dunn, Alexander W E; Hornauer, Philipp J; Ronchi, Silvia; Fiscella, Michele; Wang, Congwei; Terrigno, Marco; Jagasia, Ravi; Vértes, Petra E; Mierau, Susanna B; Paulsen, Ole; Eglen, Stephen J; Hierlemann, Andreas; Astle, Duncan E; Schröter, Manuel Homophilic wiring principles underpin neuronal network topology in vitro Journal Article BioRxiv, 2022. @article{Akarca2022, title = {Homophilic wiring principles underpin neuronal network topology in vitro}, author = {Danyal Akarca and Alexander W. E. Dunn and Philipp J. Hornauer and Silvia Ronchi and Michele Fiscella and Congwei Wang and Marco Terrigno and Ravi Jagasia and Petra E. Vértes and Susanna B. Mierau and Ole Paulsen and Stephen J. Eglen and Andreas Hierlemann and Duncan E. Astle and Manuel Schröter}, url = {https://www.biorxiv.org/content/10.1101/2022.03.09.483605v2.abstract}, doi = {https://doi.org/10.1101/2022.03.09.483605}, year = {2022}, date = {2022-12-01}, journal = {BioRxiv}, abstract = {Economic efficiency has been a popular explanation for how networks self-organize within the developing nervous system. However, the precise nature of the economic negotiations governing this putative organizational principle remains unclear. Here, we address this question further by combining large-scale electrophysiological recordings, to characterize the functional connectivity of developing neuronal networks in vitro, with a generative modeling approach capable of simulating network formation. We find that the best fitting model uses a homophilic generative wiring principle in which neurons form connections to other neurons which are spatially proximal and have similar connectivity patterns to themselves. Homophilic generative models outperform more canonical models in which neurons wire depending upon their spatial proximity either alone or in combination with the extent of their local connectivity. This homophily-based mechanism for neuronal network emergence accounts for a wide range of observations that are described, but not sufficiently explained, by traditional analyses of network topology. Using rodent and human monolayer and organoid cultures, we show that homophilic generative mechanisms can accurately recapitulate the topology of emerging cellular functional connectivity, representing an important wiring principle and determining factor of neuronal network formation in vitro.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Economic efficiency has been a popular explanation for how networks self-organize within the developing nervous system. However, the precise nature of the economic negotiations governing this putative organizational principle remains unclear. Here, we address this question further by combining large-scale electrophysiological recordings, to characterize the functional connectivity of developing neuronal networks in vitro, with a generative modeling approach capable of simulating network formation. We find that the best fitting model uses a homophilic generative wiring principle in which neurons form connections to other neurons which are spatially proximal and have similar connectivity patterns to themselves. Homophilic generative models outperform more canonical models in which neurons wire depending upon their spatial proximity either alone or in combination with the extent of their local connectivity. This homophily-based mechanism for neuronal network emergence accounts for a wide range of observations that are described, but not sufficiently explained, by traditional analyses of network topology. Using rodent and human monolayer and organoid cultures, we show that homophilic generative mechanisms can accurately recapitulate the topology of emerging cellular functional connectivity, representing an important wiring principle and determining factor of neuronal network formation in vitro. |
  | Habibollahi, Forough; Khajehnejad, Moein; Gaurav, Amitesh; Kagan, Brett Joseph Biological Neurons vs Deep Reinforcement Learning: Sample efficiency in a simulated game-world Conference 2022. @conference{Habibollahi2022, title = {Biological Neurons vs Deep Reinforcement Learning: Sample efficiency in a simulated game-world }, author = {Forough Habibollahi and Moein Khajehnejad and Amitesh Gaurav and Brett Joseph Kagan}, url = {https://openreview.net/forum?id=N5qLXpc7HQy}, year = {2022}, date = {2022-11-28}, journal = {OpenReview.net}, abstract = {How do synthetic biological systems and artificial neural networks compete in their performance in a game environment? Reinforcement learning has undergone significant advances, however remains behind biological neural intelligence in terms of sample efficiency. Yet most biological systems are significantly more complicated than most algorithms. Here we compare the inherent intelligence of in vitro biological neuronal networks to state-of-the-art deep reinforcement learning algorithms in the arcade game 'pong'. We employed DishBrain, a system that embodies in vitro neural networks with in silico computation using a high-density multielectrode array. We compared the learning curve and the performance of these biological systems against time-matched learning from DQN, A2C, and PPO algorithms. Agents were implemented in a reward-based environment of the `Pong' game. Key learning characteristics of the deep reinforcement learning agents were tested with those of the biological neuronal cultures in the same game environment. We find that even these very simple biological cultures typically outperform deep reinforcement learning systems in terms of various game performance characteristics, such as the average rally length implying a higher sample efficiency. Furthermore, the human cell cultures proved to have the overall highest relative improvement in the average number of hits in a rally when comparing the initial 5 minutes and the last 15 minutes of each designed gameplay session. }, keywords = {}, pubstate = {published}, tppubtype = {conference} } How do synthetic biological systems and artificial neural networks compete in their performance in a game environment? Reinforcement learning has undergone significant advances, however remains behind biological neural intelligence in terms of sample efficiency. Yet most biological systems are significantly more complicated than most algorithms. Here we compare the inherent intelligence of in vitro biological neuronal networks to state-of-the-art deep reinforcement learning algorithms in the arcade game 'pong'. We employed DishBrain, a system that embodies in vitro neural networks with in silico computation using a high-density multielectrode array. We compared the learning curve and the performance of these biological systems against time-matched learning from DQN, A2C, and PPO algorithms. Agents were implemented in a reward-based environment of the `Pong' game. Key learning characteristics of the deep reinforcement learning agents were tested with those of the biological neuronal cultures in the same game environment. We find that even these very simple biological cultures typically outperform deep reinforcement learning systems in terms of various game performance characteristics, such as the average rally length implying a higher sample efficiency. Furthermore, the human cell cultures proved to have the overall highest relative improvement in the average number of hits in a rally when comparing the initial 5 minutes and the last 15 minutes of each designed gameplay session. |
  | McSweeney, Danny; Gabriel, Rafael; Jin, Kang; Pang, Zhiping P; Aronow, Bruce; and Pak, ChangHui CASK loss of function differentially regulates neuronal maturation and synaptic function in human induced cortical excitatory neurons Journal Article iScience, 2022. @article{McSweeney2022b, title = {CASK loss of function differentially regulates neuronal maturation and synaptic function in human induced cortical excitatory neurons}, author = {Danny McSweeney and Rafael Gabriel and Kang Jin and Zhiping P. Pang and Bruce Aronow and and ChangHui Pak}, url = {https://www.cell.com/iscience/fulltext/S2589-0042(22)01459-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS2589004222014596%3Fshowall%3Dtrue}, doi = {https://doi.org/10.1016/j.isci.2022.105187}, year = {2022}, date = {2022-10-21}, journal = {iScience}, abstract = {Loss-of-function (LOF) mutations in CASK cause severe developmental pheno- types, including microcephaly with pontine and cerebellar hypoplasia, X-linked in- tellectual disability, and autism. Unraveling the pathological mechanisms of CASK-related disorders has been challenging owing to limited human cellular models to study the dynamic roles of this molecule during neuronal maturation and synapse development. Here, we investigate cell-autonomous functions of CASK in cortical excitatory induced neurons (iNs) generated from CASK knockout (KO) isogenic human embryonic stem cells (hESCs) using gene expression, mor- phometrics, and electrophysiology. While immature CASK KO iNs show robust neuronal outgrowth, mature CASK KO iNs display severe defects in syn- aptic transmission and synchronized network activity without compromising neuronal morphology and synapse numbers. In the developing human cortical excitatory neurons, CASK functions to promote both structural integrity and establishment of cortical excitatory neuronal networks. These results lay the foundation for future studies identifying suppressors of such phenotypes rele- vant to human patients.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Loss-of-function (LOF) mutations in CASK cause severe developmental pheno- types, including microcephaly with pontine and cerebellar hypoplasia, X-linked in- tellectual disability, and autism. Unraveling the pathological mechanisms of CASK-related disorders has been challenging owing to limited human cellular models to study the dynamic roles of this molecule during neuronal maturation and synapse development. Here, we investigate cell-autonomous functions of CASK in cortical excitatory induced neurons (iNs) generated from CASK knockout (KO) isogenic human embryonic stem cells (hESCs) using gene expression, mor- phometrics, and electrophysiology. While immature CASK KO iNs show robust neuronal outgrowth, mature CASK KO iNs display severe defects in syn- aptic transmission and synchronized network activity without compromising neuronal morphology and synapse numbers. In the developing human cortical excitatory neurons, CASK functions to promote both structural integrity and establishment of cortical excitatory neuronal networks. These results lay the foundation for future studies identifying suppressors of such phenotypes rele- vant to human patients. |
  | Kagan, Brett J; Kitchen, Andy C; Tran, Nhi T; Habibollahi, Forough; Khajehnejad, Moein; Parker, Bradyn J; Bhat, Anjali; Rollo, Ben; Razi, Adeel; Friston, Karl J In vitro neurons learn and exhibit sentience when embodied in a simulated game-world Journal Article Neuron, 2022. @article{Kagan2022, title = {In vitro neurons learn and exhibit sentience when embodied in a simulated game-world}, author = {Brett J. Kagan and Andy C. Kitchen and Nhi T. Tran and Forough Habibollahi and Moein Khajehnejad and Bradyn J. Parker and Anjali Bhat and Ben Rollo and Adeel Razi and Karl J. Friston}, url = {https://www.cell.com/neuron/fulltext/S0896-6273(22)00806-6?_returnURL=https%3A%2F%2Flinkinghub.elsevier.com%2Fretrieve%2Fpii%2FS0896627322008066%3Fshowall%3Dtrue#articleInformation}, doi = {https://doi.org/10.1016/j.neuron.2022.09.001}, year = {2022}, date = {2022-10-12}, journal = {Neuron}, abstract = {Integrating neurons into digital systems may enable performance infeasible with silicon alone. Here, we develop DishBrain, a system that harnesses the inherent adaptive computation of neurons in a structured environment. In vitro neural networks from human or rodent origins are integrated with in silico computing via a high-density multielectrode array. Through electrophysiological stimulation and recording, cultures are embedded in a simulated game-world, mimicking the arcade game ‘‘Pong.’’ Applying implications from the theory of active inference via the free energy principle, we find apparent learning within five minutes of real-time gameplay not observed in control conditions. Further experiments demonstrate the importance of closed-loop structured feedback in eliciting learning over time. Cultures display the ability to self-organize activity in a goal-directed manner in response to sparse sensory information about the consequences of their actions, which we term synthetic biological intelligence. Future applications may provide further insights into the cellular correlates of intelligence.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Integrating neurons into digital systems may enable performance infeasible with silicon alone. Here, we develop DishBrain, a system that harnesses the inherent adaptive computation of neurons in a structured environment. In vitro neural networks from human or rodent origins are integrated with in silico computing via a high-density multielectrode array. Through electrophysiological stimulation and recording, cultures are embedded in a simulated game-world, mimicking the arcade game ‘‘Pong.’’ Applying implications from the theory of active inference via the free energy principle, we find apparent learning within five minutes of real-time gameplay not observed in control conditions. Further experiments demonstrate the importance of closed-loop structured feedback in eliciting learning over time. Cultures display the ability to self-organize activity in a goal-directed manner in response to sparse sensory information about the consequences of their actions, which we term synthetic biological intelligence. Future applications may provide further insights into the cellular correlates of intelligence. |
  | Habibey, Rouhollah; Striebel, Johannes; Schmieder, Felix; Czarske, Jürgen; Busskamp, Volker Long-term morphological and functional dynamics of human stem cell-derived neuronal networks on high-density micro-electrode arrays Journal Article Frontiers in Neuroscience, 2022. @article{Habibey2022, title = {Long-term morphological and functional dynamics of human stem cell-derived neuronal networks on high-density micro-electrode arrays}, author = {Rouhollah Habibey and Johannes Striebel and Felix Schmieder and Jürgen Czarske and Volker Busskamp}, url = {https://www.frontiersin.org/articles/10.3389/fnins.2022.951964/full}, doi = {10.3389/fnins.2022.951964}, year = {2022}, date = {2022-10-04}, journal = {Frontiers in Neuroscience}, abstract = {Comprehensive electrophysiological characterizations of human induced pluripotent stem cell (hiPSC)-derived neuronal networks are essential to determine to what extent these in vitro models recapitulate the functional features of in vivo neuronal circuits. High-density micro-electrode arrays (HD-MEAs) offer non-invasive recording with the best spatial and temporal resolution possible to date. For 3 months, we tracked the morphology and activity features of developing networks derived from a transgenic hiPSC line in which neurogenesis is inducible by neurogenic transcription factor overexpression. Our morphological data revealed large-scale structural changes from homogeneously distributed neurons in the first month to the formation of neuronal clusters over time. This led to a constant shift in position of neuronal cells and clusters on HD-MEAs and corresponding changes in spatial distribution of the network activity maps. Network activity appeared as scarce action potentials (APs), evolved as local bursts with longer duration and changed to network-wide synchronized bursts with higher frequencies but shorter duration over time, resembling the emerging burst features found in the developing human brain. Instantaneous firing rate data indicated that the fraction of fast spiking neurons (150–600 Hz) increases sharply after 63 days post induction (dpi). Inhibition of glutamatergic synapses erased burst features from network activity profiles and confirmed the presence of mature excitatory neurotransmission. The application of GABAergic receptor antagonists profoundly changed the bursting profile of the network at 120 dpi. This indicated a GABAergic switch from excitatory to inhibitory neurotransmission during circuit development and maturation. Our results suggested that an emerging GABAergic system at older culture ages is involved in regulating spontaneous network bursts. In conclusion, our data showed that long-term and continuous microscopy and electrophysiology readouts are crucial for a meaningful characterization of morphological and functional maturation in stem cell-derived human networks. Most importantly, assessing the level and duration of functional maturation is key to subject these human neuronal circuits on HD-MEAs for basic and biomedical applications.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Comprehensive electrophysiological characterizations of human induced pluripotent stem cell (hiPSC)-derived neuronal networks are essential to determine to what extent these in vitro models recapitulate the functional features of in vivo neuronal circuits. High-density micro-electrode arrays (HD-MEAs) offer non-invasive recording with the best spatial and temporal resolution possible to date. For 3 months, we tracked the morphology and activity features of developing networks derived from a transgenic hiPSC line in which neurogenesis is inducible by neurogenic transcription factor overexpression. Our morphological data revealed large-scale structural changes from homogeneously distributed neurons in the first month to the formation of neuronal clusters over time. This led to a constant shift in position of neuronal cells and clusters on HD-MEAs and corresponding changes in spatial distribution of the network activity maps. Network activity appeared as scarce action potentials (APs), evolved as local bursts with longer duration and changed to network-wide synchronized bursts with higher frequencies but shorter duration over time, resembling the emerging burst features found in the developing human brain. Instantaneous firing rate data indicated that the fraction of fast spiking neurons (150–600 Hz) increases sharply after 63 days post induction (dpi). Inhibition of glutamatergic synapses erased burst features from network activity profiles and confirmed the presence of mature excitatory neurotransmission. The application of GABAergic receptor antagonists profoundly changed the bursting profile of the network at 120 dpi. This indicated a GABAergic switch from excitatory to inhibitory neurotransmission during circuit development and maturation. Our results suggested that an emerging GABAergic system at older culture ages is involved in regulating spontaneous network bursts. In conclusion, our data showed that long-term and continuous microscopy and electrophysiology readouts are crucial for a meaningful characterization of morphological and functional maturation in stem cell-derived human networks. Most importantly, assessing the level and duration of functional maturation is key to subject these human neuronal circuits on HD-MEAs for basic and biomedical applications. |
  | Lee, Jihyun; Gänswein, Tobias; Ulusan, Hasan; Emmenegger, Vishalini; Saguner, Ardan M; Duru, Firat; and Hierlemann, Andreas Repeated and On-Demand Intracellular Recordings of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells Journal Article ACS Sensors, 2022. @article{Lee2022, title = {Repeated and On-Demand Intracellular Recordings of Cardiomyocytes Derived from Human Induced Pluripotent Stem Cells}, author = {Jihyun Lee and Tobias Gänswein and Hasan Ulusan and Vishalini Emmenegger and Ardan M. Saguner and Firat Duru and and Andreas Hierlemann}, url = {https://pubs.acs.org/doi/10.1021/acssensors.2c01678}, doi = {https://doi.org/10.1021/acssensors.2c01678}, year = {2022}, date = {2022-09-27}, journal = {ACS Sensors}, abstract = {Pharmaceutical compounds may have cardiotoxic properties, triggering potentially life-threatening arrhythmi- as. To investigate proarrhythmic effects of drugs, the patch clamp technique has been used as the gold standard for charac- terizing the electrophysiology of cardiomyocytes in vitro. However, the applicability of this technology for drug screening is limited, as it is complex to use and features low throughput. Recent studies have demonstrated that 3D-nanostructured electrodes enable to obtain intracellular signals from many cardiomyocytes in parallel; however, the tedious electrode fab- rication and limited measurement duration still remain major issues for cardiotoxicity testing. Here, we demonstrate how porous Pt-black electrodes, arranged in high-density microelectrode arrays, can be used to record intracellular-like signals of cardiomyocytes at large-scale repeatedly over an extended period of time. The developed technique, which yields highly parallelized electroporations by using stimulation voltages around 1 Volt peak-to-peak amplitude, enabled intracellular-like recordings at high success rates without causing significant alteration in key electrophysiological features. In a proof of concept study, we investigated electrophysiological modulations induced by two clinically applied drugs, nifedipine and quinidine. As the obtained results were in good agreement with previously published data, we are confident that the devel- oped technique has the potential to be routinely used in in vitro platforms for cardiotoxicity screening.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Pharmaceutical compounds may have cardiotoxic properties, triggering potentially life-threatening arrhythmi- as. To investigate proarrhythmic effects of drugs, the patch clamp technique has been used as the gold standard for charac- terizing the electrophysiology of cardiomyocytes in vitro. However, the applicability of this technology for drug screening is limited, as it is complex to use and features low throughput. Recent studies have demonstrated that 3D-nanostructured electrodes enable to obtain intracellular signals from many cardiomyocytes in parallel; however, the tedious electrode fab- rication and limited measurement duration still remain major issues for cardiotoxicity testing. Here, we demonstrate how porous Pt-black electrodes, arranged in high-density microelectrode arrays, can be used to record intracellular-like signals of cardiomyocytes at large-scale repeatedly over an extended period of time. The developed technique, which yields highly parallelized electroporations by using stimulation voltages around 1 Volt peak-to-peak amplitude, enabled intracellular-like recordings at high success rates without causing significant alteration in key electrophysiological features. In a proof of concept study, we investigated electrophysiological modulations induced by two clinically applied drugs, nifedipine and quinidine. As the obtained results were in good agreement with previously published data, we are confident that the devel- oped technique has the potential to be routinely used in in vitro platforms for cardiotoxicity screening. |
  | Al-Absi, Abdel-Rahman; Thambiappaa, Sakeerthi Kethees; Khanc, Ahmad Raza; Glerup, Simon; Sanchez, Connie; Landau, Anne M; Nyengaard, Jens R Molecular and Cellular Neuroscience, 2022. @article{Al-Absi2022, title = {Df(h22q11)/+ mouse model exhibits reduced binding levels of GABAA receptors and structural and functional dysregulation in the inhibitory and excitatory networks of hippocampus}, author = {Abdel-Rahman Al-Absi and Sakeerthi Kethees Thambiappaa and Ahmad Raza Khanc and Simon Glerup and Connie Sanchez and Anne M. Landau and Jens R. Nyengaard}, url = {https://www.sciencedirect.com/science/article/pii/S1044743122000756?via%3Dihub}, doi = {https://doi.org/10.1016/j.mcn.2022.103769}, year = {2022}, date = {2022-08-18}, journal = {Molecular and Cellular Neuroscience}, abstract = {The 22q11.2 hemizygous deletion confers high risk for multiple neurodevelopmental disorders. Inhibitory signaling, largely regulated through GABAA receptors, is suggested to serve a multitude of brain functions that are disrupted in the 22q11.2 deletion syndrome. We investigated the putative deficit of GABAA receptors and the potential substrates contributing to the inhibitory and excitatory dysregulations in hippocampal networks of the Df(h22q11)/+ mouse model of the 22q11.2 hemizygous deletion. The Df(h22q11)/+ mice exhibited impairments in several hippocampus-related functional domains, represented by impaired spatial memory and sensory gating functions. Autoradiography using the [3H]muscimol tracer revealed a significant reduction in GABAA receptor binding in the CA1 and CA3 subregions, together with a loss of GAD67+ interneurons in CA1 of Df(h22q11)/+ mice. Furthermore, electro- physiology recordings exhibited significantly higher neuronal activity in CA3, in response to the GABAA receptor antagonist, bicuculline, as compared with wild type mice. Density and volume of dendritic spines in pyramidal neurons were reduced and Sholl analysis also showed a reduction in the complexity of basal dendritic tree in CA1 and CA3 subregions of Df(h22q11)/+ mice. Overall, our findings demonstrate that hemizygous deletion in the 22q11.2 locus leads to dysregulations in the inhibitory circuits, involving reduced binding levels of GABAA receptors, in addition to functional and structural modulations of the excitatory networks of hippocampus.}, keywords = {}, pubstate = {published}, tppubtype = {article} } The 22q11.2 hemizygous deletion confers high risk for multiple neurodevelopmental disorders. Inhibitory signaling, largely regulated through GABAA receptors, is suggested to serve a multitude of brain functions that are disrupted in the 22q11.2 deletion syndrome. We investigated the putative deficit of GABAA receptors and the potential substrates contributing to the inhibitory and excitatory dysregulations in hippocampal networks of the Df(h22q11)/+ mouse model of the 22q11.2 hemizygous deletion. The Df(h22q11)/+ mice exhibited impairments in several hippocampus-related functional domains, represented by impaired spatial memory and sensory gating functions. Autoradiography using the [3H]muscimol tracer revealed a significant reduction in GABAA receptor binding in the CA1 and CA3 subregions, together with a loss of GAD67+ interneurons in CA1 of Df(h22q11)/+ mice. Furthermore, electro- physiology recordings exhibited significantly higher neuronal activity in CA3, in response to the GABAA receptor antagonist, bicuculline, as compared with wild type mice. Density and volume of dendritic spines in pyramidal neurons were reduced and Sholl analysis also showed a reduction in the complexity of basal dendritic tree in CA1 and CA3 subregions of Df(h22q11)/+ mice. Overall, our findings demonstrate that hemizygous deletion in the 22q11.2 locus leads to dysregulations in the inhibitory circuits, involving reduced binding levels of GABAA receptors, in addition to functional and structural modulations of the excitatory networks of hippocampus. |
  | Buccino Alessio Paolo; Damart, Tanguy; Bartram Julian; Mandge Darshan; Xue Xiaohan; Zbili Mickael; Gänswein Tobias; Jaquier Aurélien; Emmenegger Vishalini; Markram Henry; Hierlemann Andreas; Van Geit Werner. A multi-modal fitting approach to construct single-neuron models with patch clamp and high-density microelectrode arrays Journal Article bioRxiv, 2022. @article{Buccino2022, title = {A multi-modal fitting approach to construct single-neuron models with patch clamp and high-density microelectrode arrays}, author = {Buccino, Alessio Paolo; Damart, Tanguy; Bartram, Julian; Mandge, Darshan; Xue, Xiaohan; Zbili, Mickael; Gänswein, Tobias; Jaquier, Aurélien; Emmenegger, Vishalini; Markram, Henry; Hierlemann, Andreas; Van Geit, Werner.}, doi = {https://doi.org/10.1101/2022.08.03.502468}, year = {2022}, date = {2022-08-11}, journal = {bioRxiv}, abstract = {In computational neuroscience, multicompartment models are among the most biophysically realistic representations of single neurons. Constructing such models usually involves the use of the patch-clamp technique to record somatic voltage signals under different experimental conditions. The experimental data are then used to fit the many parameters of the model. While patching of the soma is currently the gold-standard approach to build multicompartment models, several studies have also evidenced a richness of dynamics in dendritic and axonal sections. Recording from the soma alone makes it hard to observe and correctly parameterize the activity of non-somatic compartments. In order to provide a richer set of data as input to multicompartment models, we here investigate the combination of somatic patch-clamp recordings with recordings of high-density micro-electrode arrays (HD-MEAs). HD-MEAs enable the observation of extracellular potentials and neural activity of neuronal compartments at sub-cellular resolution. In this work, we introduce a novel framework to combine patch-clamp and HD-MEA data to construct multicompartment models. We first validate our method on a ground-truth model with known parameters and show that the use of features extracted from extracellular signals, in addition to intracellular ones, yields models enabling better fits than using intracellular features alone. We also demonstrate our procedure using experimental data by constructing cell models from in vitro cell cultures. The proposed multi-modal fitting procedure has the potential to augment the modeling efforts of the computational neuroscience community and to provide the field with neuronal models that are more realistic and can be better validated. Author Summary Multicompartment models are one of the most biophysically detailed representations of single neurons. The vast majority of these models are built using experimental data from somatic recordings. However, neurons are much more than just their soma and one needs recordings from distal neurites to build an accurate model. In this article, we combine the patch-clamp technique with extracellular high-density microelectrode arrays (HD-MEAs) to compensate this shortcoming. In fact, HD-MEAs readouts allow one to record the neuronal signal in the entire axonal arbor. We show that the proposed multi-modal strategy is superior to the use of patch clamp alone using an existing model as ground-truth. Finally, we show an application of this strategy on experimental data from cultured neurons.}, keywords = {}, pubstate = {published}, tppubtype = {article} } In computational neuroscience, multicompartment models are among the most biophysically realistic representations of single neurons. Constructing such models usually involves the use of the patch-clamp technique to record somatic voltage signals under different experimental conditions. The experimental data are then used to fit the many parameters of the model. While patching of the soma is currently the gold-standard approach to build multicompartment models, several studies have also evidenced a richness of dynamics in dendritic and axonal sections. Recording from the soma alone makes it hard to observe and correctly parameterize the activity of non-somatic compartments. In order to provide a richer set of data as input to multicompartment models, we here investigate the combination of somatic patch-clamp recordings with recordings of high-density micro-electrode arrays (HD-MEAs). HD-MEAs enable the observation of extracellular potentials and neural activity of neuronal compartments at sub-cellular resolution. In this work, we introduce a novel framework to combine patch-clamp and HD-MEA data to construct multicompartment models. We first validate our method on a ground-truth model with known parameters and show that the use of features extracted from extracellular signals, in addition to intracellular ones, yields models enabling better fits than using intracellular features alone. We also demonstrate our procedure using experimental data by constructing cell models from in vitro cell cultures. The proposed multi-modal fitting procedure has the potential to augment the modeling efforts of the computational neuroscience community and to provide the field with neuronal models that are more realistic and can be better validated. Author Summary Multicompartment models are one of the most biophysically detailed representations of single neurons. The vast majority of these models are built using experimental data from somatic recordings. However, neurons are much more than just their soma and one needs recordings from distal neurites to build an accurate model. In this article, we combine the patch-clamp technique with extracellular high-density microelectrode arrays (HD-MEAs) to compensate this shortcoming. In fact, HD-MEAs readouts allow one to record the neuronal signal in the entire axonal arbor. We show that the proposed multi-modal strategy is superior to the use of patch clamp alone using an existing model as ground-truth. Finally, we show an application of this strategy on experimental data from cultured neurons. |
  | Xue, Xiaohan; Buccino, Alessio Paolo; Kumar, Sreedhar Saseendran; Hierlemann, Andreas; Bartram, Julian Inferring monosynaptic connections from paired dendritic spine Ca2+ imaging and large-scale recording of extracellular spiking Journal Article Journal of Neural Engineering, 2022. @article{Xue2022b, title = {Inferring monosynaptic connections from paired dendritic spine Ca2+ imaging and large-scale recording of extracellular spiking}, author = {Xiaohan Xue and Alessio Paolo Buccino and Sreedhar Saseendran Kumar and Andreas Hierlemann and Julian Bartram}, doi = {https://doi.org/10.1088/1741-2552/ac8765}, year = {2022}, date = {2022-08-11}, journal = {Journal of Neural Engineering}, abstract = {Techniques to identify monosynaptic connections between neurons have been vital for neuroscience research, facilitating important advancements concerning network topology, synaptic plasticity, and synaptic integration, among others. Here, we introduce a novel approach to identify and monitor monosynaptic connections using high-resolution dendritic spine Ca2+ imaging combined with simultaneous large-scale recording of extracellular electrical activity by means of high-density microelectrode arrays (HD-MEAs). We introduce an easily adoptable analysis pipeline that associates the imaged spine with its presynaptic unit and test it on in vitro recordings. The method is further validated and optimized by simulating synaptically-evoked spine Ca2+ transients based on measured spike trains in order to obtain simulated ground-truth connections. The proposed approach offers unique advantages as i) it can be used to identify monosynaptic connections with an accurate localization of the synapse within the dendritic tree, ii) it provides precise information of presynaptic spiking, and iii) postsynaptic spine Ca2+ signals and, finally, iv) the non-invasive nature of the proposed method allows for long-term measurements. The analysis toolkit together with the rich data sets that were acquired are made publicly available for further exploration by the research community.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Techniques to identify monosynaptic connections between neurons have been vital for neuroscience research, facilitating important advancements concerning network topology, synaptic plasticity, and synaptic integration, among others. Here, we introduce a novel approach to identify and monitor monosynaptic connections using high-resolution dendritic spine Ca2+ imaging combined with simultaneous large-scale recording of extracellular electrical activity by means of high-density microelectrode arrays (HD-MEAs). We introduce an easily adoptable analysis pipeline that associates the imaged spine with its presynaptic unit and test it on in vitro recordings. The method is further validated and optimized by simulating synaptically-evoked spine Ca2+ transients based on measured spike trains in order to obtain simulated ground-truth connections. The proposed approach offers unique advantages as i) it can be used to identify monosynaptic connections with an accurate localization of the synapse within the dendritic tree, ii) it provides precise information of presynaptic spiking, and iii) postsynaptic spine Ca2+ signals and, finally, iv) the non-invasive nature of the proposed method allows for long-term measurements. The analysis toolkit together with the rich data sets that were acquired are made publicly available for further exploration by the research community. |
  | Sharf Tal; Molen, Tjitse; Glasauer Stella; Guzman Elmer; Buccino Alessio; Luna Gabriel; Cheng Zhuowei; Audouard Morgane; Ranasinghe Kamalini; Kudo Kiwamu; Nagarajan Srikantan; Tovar Kenneth; Petzold Linda; Hierlemann Andreas; Hansma Paul; ; Kosik, Kenneth; Functional neuronal circuitry and oscillatory dynamics in human brain organoids Journal Article Nature Communications, 2022. @article{Sharf2022, title = {Functional neuronal circuitry and oscillatory dynamics in human brain organoids}, author = {Sharf, Tal; Molen, Tjitse; Glasauer, Stella; Guzman, Elmer; Buccino, Alessio; Luna, Gabriel; Cheng, Zhuowei; Audouard, Morgane; Ranasinghe, Kamalini; Kudo, Kiwamu; Nagarajan, Srikantan; Tovar, Kenneth; Petzold, Linda; Hierlemann, Andreas; Hansma, Paul; and Kosik, Kenneth; }, doi = {https://doi.org/10.1038/s41467-022-32115-4}, year = {2022}, date = {2022-07-29}, journal = {Nature Communications}, abstract = {Human brain organoids replicate much of the cellular diversity and developmental anatomy of the human brain. However, the physiology of neuronal circuits within organoids remains under-explored. With high-density CMOS microelectrode arrays and shank electrodes, we captured spontaneous extracellular activity from brain organoids derived from human induced pluripotent stem cells. We inferred functional connectivity from spike timing, revealing a large number of weak connections within a skeleton of significantly fewer strong connections. A benzodiazepine increased the uniformity of firing patterns and decreased the relative fraction of weakly connected edges. Our analysis of the local field potential demonstrate that brain organoids contain neuronal assemblies of sufficient size and functional connectivity to co-activate and generate field potentials from their collective transmembrane currents that phase-lock to spiking activity. These results point to the potential of brain organoids for the study of neuropsychiatric diseases, drug action, and the effects of external stimuli upon neuronal networks.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Human brain organoids replicate much of the cellular diversity and developmental anatomy of the human brain. However, the physiology of neuronal circuits within organoids remains under-explored. With high-density CMOS microelectrode arrays and shank electrodes, we captured spontaneous extracellular activity from brain organoids derived from human induced pluripotent stem cells. We inferred functional connectivity from spike timing, revealing a large number of weak connections within a skeleton of significantly fewer strong connections. A benzodiazepine increased the uniformity of firing patterns and decreased the relative fraction of weakly connected edges. Our analysis of the local field potential demonstrate that brain organoids contain neuronal assemblies of sufficient size and functional connectivity to co-activate and generate field potentials from their collective transmembrane currents that phase-lock to spiking activity. These results point to the potential of brain organoids for the study of neuropsychiatric diseases, drug action, and the effects of external stimuli upon neuronal networks. |
  | Idrees, Saad; Baumann, Matthias-Philipp; Korympidou, Maria M; Schubert, Timm; Kling, Alexandra; Franke, Katrin; Hafed, Ziad M; Franke, Felix; Münch, Thomas A Suppression without inhibition: how retinal computation contributes to saccadic suppression Journal Article Communications Biology, 2022. @article{Idrees2022, title = {Suppression without inhibition: how retinal computation contributes to saccadic suppression}, author = {Saad Idrees and Matthias-Philipp Baumann and Maria M. Korympidou and Timm Schubert and Alexandra Kling and Katrin Franke and Ziad M. Hafed and Felix Franke and Thomas A. Münch }, url = {https://www.nature.com/articles/s42003-022-03526-2}, year = {2022}, date = {2022-07-12}, journal = {Communications Biology}, abstract = {Visual perception remains stable across saccadic eye movements, despite the concurrent strongly disruptive visual flow. This stability is partially associated with a reduction in visual sensitivity, known as saccadic suppression, which already starts in the retina with reduced ganglion cell sensitivity. However, the retinal circuit mechanisms giving rise to such sup- pression remain unknown. Here, we describe these mechanisms using electrophysiology in mouse, pig, and macaque retina, 2-photon calcium imaging, computational modeling, and human psychophysics. We find that sequential stimuli, like those that naturally occur during saccades, trigger three independent suppressive mechanisms in the retina. The main mechanism is triggered by contrast-reversing sequential stimuli and originates within the receptive field center of ganglion cells. It does not involve inhibition or other known sup- pressive mechanisms like saturation or adaptation. Instead, it relies on temporal filtering of the inherently slow response of cone photoreceptors coupled with downstream non- linearities. Two further mechanisms of suppression are present predominantly in ON ganglion cells and originate in the receptive field surround, highlighting another disparity between ON and OFF ganglion cells. The mechanisms uncovered here likely play a role in shaping the retinal output following eye movements and other natural viewing conditions where sequential stimulation is ubiquitous.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Visual perception remains stable across saccadic eye movements, despite the concurrent strongly disruptive visual flow. This stability is partially associated with a reduction in visual sensitivity, known as saccadic suppression, which already starts in the retina with reduced ganglion cell sensitivity. However, the retinal circuit mechanisms giving rise to such sup- pression remain unknown. Here, we describe these mechanisms using electrophysiology in mouse, pig, and macaque retina, 2-photon calcium imaging, computational modeling, and human psychophysics. We find that sequential stimuli, like those that naturally occur during saccades, trigger three independent suppressive mechanisms in the retina. The main mechanism is triggered by contrast-reversing sequential stimuli and originates within the receptive field center of ganglion cells. It does not involve inhibition or other known sup- pressive mechanisms like saturation or adaptation. Instead, it relies on temporal filtering of the inherently slow response of cone photoreceptors coupled with downstream non- linearities. Two further mechanisms of suppression are present predominantly in ON ganglion cells and originate in the receptive field surround, highlighting another disparity between ON and OFF ganglion cells. The mechanisms uncovered here likely play a role in shaping the retinal output following eye movements and other natural viewing conditions where sequential stimulation is ubiquitous. |
  | Schröter Manuel; Wang, Congwei; Terrigno Marco; Hornauer Philipp; Huang Ziqiang; Jagasia Ravi; Hierlemann Andreas Functional imaging of brain organoids using high-density microelectrode arrays Journal Article MRS Bulletin, 2022. @article{Schroter2022, title = {Functional imaging of brain organoids using high-density microelectrode arrays}, author = {Schröter, Manuel; Wang, Congwei; Terrigno, Marco; Hornauer, Philipp; Huang, Ziqiang; Jagasia, Ravi; Hierlemann, Andreas}, url = {https://link.springer.com/article/10.1557/s43577-022-00282-w}, year = {2022}, date = {2022-06-30}, journal = {MRS Bulletin}, abstract = {Studies have provided evidence that human cerebral organoids (hCOs) recapitulate fundamental milestones of early brain development, but many important questions regarding their functionality and electrophysiological properties persist. High-density microelectrode arrays (HD-MEAs) represent an attractive analysis platform to perform functional studies of neuronal networks at the cellular and network scale. Here, we use HD-MEAs to derive large-scale electrophysiological recordings from sliced hCOs. We record the activity of hCO slices over several weeks and probe observed neuronal dynamics pharmacologically. Moreover, we present results on how the obtained recordings can be spike-sorted and subsequently studied across scales. For example, we show how to track single neurons across several days on the HD-MEA and how to infer axonal action potential velocities. We also infer putative functional connectivity from hCO recordings. The introduced methodology will contribute to a better understanding of developing neuronal networks in brain organoids and provide new means for their functional characterization.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Studies have provided evidence that human cerebral organoids (hCOs) recapitulate fundamental milestones of early brain development, but many important questions regarding their functionality and electrophysiological properties persist. High-density microelectrode arrays (HD-MEAs) represent an attractive analysis platform to perform functional studies of neuronal networks at the cellular and network scale. Here, we use HD-MEAs to derive large-scale electrophysiological recordings from sliced hCOs. We record the activity of hCO slices over several weeks and probe observed neuronal dynamics pharmacologically. Moreover, we present results on how the obtained recordings can be spike-sorted and subsequently studied across scales. For example, we show how to track single neurons across several days on the HD-MEA and how to infer axonal action potential velocities. We also infer putative functional connectivity from hCO recordings. The introduced methodology will contribute to a better understanding of developing neuronal networks in brain organoids and provide new means for their functional characterization. |
  | Xu, Jax H; Yao, Yao; Yao, Fenyong; Chen, Jiehui; Li, Meishi; Xianfa, ; Yang, ; Li, Sheng; Lu, Fangru; Hu, Ping; He, Shuijin; Peng, Guangdun; Jing, Naihe Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells Journal Article BioRxiv, 2022. @article{Xu2022, title = {Generation of functional posterior spinal motor neurons from hPSCs-derived human spinal cord neural progenitor cells}, author = {Jax H. Xu and Yao Yao and Fenyong Yao and Jiehui Chen and Meishi Li and Xianfa and Yang and Sheng Li and Fangru Lu and Ping Hu and Shuijin He and Guangdun Peng and Naihe Jing}, url = {https://www.biorxiv.org/content/10.1101/2022.06.26.495599v1}, doi = {https://doi.org/10.1101/2022.06.26.495599}, year = {2022}, date = {2022-06-27}, journal = {BioRxiv}, abstract = {Spinal motor neurons deficiency results in a series of devastating disorders such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA) and spinal cord injury (SCI). These disorders are currently incurable, while human pluripotent stem cells (hPSCs)-derived spinal motor neurons are promising but suffered from low-efficiency, functional immaturity and lacks of posterior cellular identity. In this study, we have established human spinal cord neural progenitor cells (hSCNPCs) via hPSCs differentiated neuromesodermal progenitors (NMPs) and demonstrated the hSCNPCs can be continuously expanded up to 40 passages. hSCNPCs can be rapidly differentiated into posterior spinal motor neurons with high efficiency. The functional maturity has been examined in detail. Moreover, a co-culture scheme which is compatible for both neural and muscular differentiation is developed to mimic the neuromuscular junction (NMJ) formation in vitro. Together, these studies highlight the potential avenues for generating clinically relevant spinal motor neurons and modeling neuromuscular diseases through our defined hSCNPCs.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Spinal motor neurons deficiency results in a series of devastating disorders such as amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA) and spinal cord injury (SCI). These disorders are currently incurable, while human pluripotent stem cells (hPSCs)-derived spinal motor neurons are promising but suffered from low-efficiency, functional immaturity and lacks of posterior cellular identity. In this study, we have established human spinal cord neural progenitor cells (hSCNPCs) via hPSCs differentiated neuromesodermal progenitors (NMPs) and demonstrated the hSCNPCs can be continuously expanded up to 40 passages. hSCNPCs can be rapidly differentiated into posterior spinal motor neurons with high efficiency. The functional maturity has been examined in detail. Moreover, a co-culture scheme which is compatible for both neural and muscular differentiation is developed to mimic the neuromuscular junction (NMJ) formation in vitro. Together, these studies highlight the potential avenues for generating clinically relevant spinal motor neurons and modeling neuromuscular diseases through our defined hSCNPCs. |
  | Sommer Daniel ; Rajkumar, Sandeep; Seidel Mira; Aly Amr; Ludolph Albert; Ho Ritchie; Boeckers Tobias; Catanese Alberto. Aging-Dependent Altered Transcriptional Programs Underlie Activity Impairments in Human C9orf72-Mutant Motor Neurons Journal Article Frontiers in Molecular Neuroscience, 2022. @article{Sommer2022, title = {Aging-Dependent Altered Transcriptional Programs Underlie Activity Impairments in Human C9orf72-Mutant Motor Neurons}, author = {Sommer, Daniel ; Rajkumar, Sandeep; Seidel, Mira; Aly, Amr; Ludolph, Albert; Ho, Ritchie; Boeckers, Tobias; Catanese, Alberto.}, url = {https://www.frontiersin.org/articles/10.3389/fnmol.2022.894230/full}, year = {2022}, date = {2022-06-14}, journal = {Frontiers in Molecular Neuroscience}, abstract = {Amyotrophic Lateral Sclerosis (ALS) is an incurable neurodegenerative disease characterized by dysfunction and loss of upper and lower motor neurons (MN). Despite several studies identifying drastic alterations affecting synaptic composition and functionality in different experimental models, the specific contribution of impaired activity to the neurodegenerative processes observed in ALS-related MN remains controversial. In particular, contrasting lines of evidence have shown both hyper- as well as hypoexcitability as driving pathomechanisms characterizing this specific neuronal population. In this study, we combined high definition multielectrode array (HD-MEA) techniques with transcriptomic analysis to longitudinally monitor and untangle the activity-dependent alterations arising in human C9orf72-mutant MN. We found a time-dependent reduction of neuronal activity in ALSC9orf72 cultures occurring as synaptic contacts undergo maturation and matched by a significant loss of mutant MN upon aging. Notably, ALS-related neurons displayed reduced network synchronicity most pronounced at later stages of culture, suggesting synaptic imbalance. In concordance with the HD-MEA data, transcriptomic analysis revealed an early up-regulation of synaptic terms in ALSC9orf72 MN, whose expression was decreased in aged cultures. In addition, treatment of older mutant cells with Apamin, a K+ channel blocker previously shown to be neuroprotective in ALS, rescued the time-dependent loss of firing properties observed in ALSC9orf72 MN as well as the expression of maturity-related synaptic genes. All in all, this study broadens the understanding of how impaired synaptic activity contributes to MN degeneration in ALS by correlating electrophysiological alterations to aging-dependent transcriptional programs. }, keywords = {}, pubstate = {published}, tppubtype = {article} } Amyotrophic Lateral Sclerosis (ALS) is an incurable neurodegenerative disease characterized by dysfunction and loss of upper and lower motor neurons (MN). Despite several studies identifying drastic alterations affecting synaptic composition and functionality in different experimental models, the specific contribution of impaired activity to the neurodegenerative processes observed in ALS-related MN remains controversial. In particular, contrasting lines of evidence have shown both hyper- as well as hypoexcitability as driving pathomechanisms characterizing this specific neuronal population. In this study, we combined high definition multielectrode array (HD-MEA) techniques with transcriptomic analysis to longitudinally monitor and untangle the activity-dependent alterations arising in human C9orf72-mutant MN. We found a time-dependent reduction of neuronal activity in ALSC9orf72 cultures occurring as synaptic contacts undergo maturation and matched by a significant loss of mutant MN upon aging. Notably, ALS-related neurons displayed reduced network synchronicity most pronounced at later stages of culture, suggesting synaptic imbalance. In concordance with the HD-MEA data, transcriptomic analysis revealed an early up-regulation of synaptic terms in ALSC9orf72 MN, whose expression was decreased in aged cultures. In addition, treatment of older mutant cells with Apamin, a K+ channel blocker previously shown to be neuroprotective in ALS, rescued the time-dependent loss of firing properties observed in ALSC9orf72 MN as well as the expression of maturity-related synaptic genes. All in all, this study broadens the understanding of how impaired synaptic activity contributes to MN degeneration in ALS by correlating electrophysiological alterations to aging-dependent transcriptional programs. |
  | Cheng, Zhuowei; Guzman, Elmer; van der Molen, Tjitse; Sharf, Tal; Hansma, Paul K; Kosik, Kenneth S; Petzold, Linda; Tovar, Kenneth R Automated detection of extracellular action potentials from single neurons Journal Article bioRxiv, 2022. @article{Cheng2023, title = {Automated detection of extracellular action potentials from single neurons}, author = {Zhuowei Cheng and Elmer Guzman and Tjitse van der Molen and Tal Sharf and Paul K. Hansma and Kenneth S Kosik and Linda Petzold and Kenneth R Tovar}, url = {https://www.biorxiv.org/content/10.1101/2022.06.06.494896v1.abstract}, doi = {https://doi.org/10.1101/2022.06.06.494896}, year = {2022}, date = {2022-06-06}, journal = {bioRxiv}, abstract = {Multi-electrode arrays (MEAs) non-invasively record extracellular action potentials (eAPs, also known as spikes) from hundreds of neurons simultaneously. However, because extracellular electrodes sample from the local electrical field, each electrode can detect eAPs from multiple nearby neurons. Interpreting spike trains at individual electrodes of high-density arrays requires spike sorting, a computational process which groups eAPs from single ’units’ based on assumptions of how spike waveforms correlate with different neuronal sources. Additionally, when experimental conditions result in changes to eAP waveforms, spike sorting routines may have difficulty correlating eAPs from multiple neurons at single electrodes before and after such waveform changes. We present here a novel, empirical method for unambiguously isolating eAPs from individual, uniquely identifiable neurons, based on automated multi- point detection of action potential propagation. This method is insensitive to changes in eAP waveform morphology because it makes no assumptions about the relationship between spike waveform and neuronal source. Our algorithm for automated detection of action potential propagation produces a ’fingerprint’ that uniquely identifies those spikes from each neuron. By unambiguously isolating eAPs from multiple neurons in each recording, on a range of platforms and experimental preparations, our method now enables high-content screening with contemporary MEAs. We outline the limitations and strengths of propagation-based isolation of eAPs from single neurons and propose how our automated method complements spike sorting and could be adapted to in vivo use.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Multi-electrode arrays (MEAs) non-invasively record extracellular action potentials (eAPs, also known as spikes) from hundreds of neurons simultaneously. However, because extracellular electrodes sample from the local electrical field, each electrode can detect eAPs from multiple nearby neurons. Interpreting spike trains at individual electrodes of high-density arrays requires spike sorting, a computational process which groups eAPs from single ’units’ based on assumptions of how spike waveforms correlate with different neuronal sources. Additionally, when experimental conditions result in changes to eAP waveforms, spike sorting routines may have difficulty correlating eAPs from multiple neurons at single electrodes before and after such waveform changes. We present here a novel, empirical method for unambiguously isolating eAPs from individual, uniquely identifiable neurons, based on automated multi- point detection of action potential propagation. This method is insensitive to changes in eAP waveform morphology because it makes no assumptions about the relationship between spike waveform and neuronal source. Our algorithm for automated detection of action potential propagation produces a ’fingerprint’ that uniquely identifies those spikes from each neuron. By unambiguously isolating eAPs from multiple neurons in each recording, on a range of platforms and experimental preparations, our method now enables high-content screening with contemporary MEAs. We outline the limitations and strengths of propagation-based isolation of eAPs from single neurons and propose how our automated method complements spike sorting and could be adapted to in vivo use. |
  | Noh Seungmin ; Jeon, Sungwoong; Kim Eunhee; Oh Untaek; Park Danbi; Park Sun Hwa; Kim Sung Won; Pané Salvador; Nelson Bradley; Kim Jin-young; Choi Hongsoo; A Biodegradable Magnetic Microrobot Based on Gelatin Methacrylate for Precise Delivery of Stem Cells with Mass Production Capability Journal Article Small, 2022. @article{Noh2022, title = {A Biodegradable Magnetic Microrobot Based on Gelatin Methacrylate for Precise Delivery of Stem Cells with Mass Production Capability}, author = {Noh, Seungmin ; Jeon, Sungwoong; Kim, Eunhee; Oh, Untaek; Park, Danbi; Park, Sun Hwa; Kim, Sung Won; Pané, Salvador; Nelson, Bradley; Kim, Jin-young; Choi, Hongsoo;}, url = {https://onlinelibrary.wiley.com/doi/10.1002/smll.202107888}, doi = {10.1002/smll.202107888}, year = {2022}, date = {2022-05-23}, journal = {Small}, abstract = {A great deal of research has focused on small-scale robots for biomedical applications and minimally invasive delivery of therapeutics (e.g., cells, drugs, and genes) to a target area. Conventional fabrication methods, such as two-photon polymerization, can be used to build sophisticated micro- and nanorobots, but the long fabrication cycle for a single microrobot has limited its practical use. This study proposes a biodegradable spherical gelatin methacrylate (GelMA) microrobot for mass production in a microfluidic channel. The proposed microrobot is fabricated in a flow-focusing droplet generator by shearing a mixture of GelMA, photoinitiator, and superparamagnetic iron oxide nanoparticles (SPIONs) with a mixture of oil and surfactant. Human nasal turbinate stem cells (hNTSCs) are loaded on the GelMA microrobot, and the hNTSC-loaded microrobot shows precise rolling motion in response to an external rotating magnetic field. The microrobot is enzymatically degraded by collagenase, and released hNTSCs are proliferated and differentiated into neuronal cells. In addition, the feasibility of the GelMA microrobot as a cell therapeutic delivery system is investigated by measuring electrophysiological activity on a multielectrode array. Such a versatile and fully biodegradable microrobot has the potential for targeted stem cell delivery, proliferation, and differentiation for stem cell-based therapy.}, keywords = {}, pubstate = {published}, tppubtype = {article} } A great deal of research has focused on small-scale robots for biomedical applications and minimally invasive delivery of therapeutics (e.g., cells, drugs, and genes) to a target area. Conventional fabrication methods, such as two-photon polymerization, can be used to build sophisticated micro- and nanorobots, but the long fabrication cycle for a single microrobot has limited its practical use. This study proposes a biodegradable spherical gelatin methacrylate (GelMA) microrobot for mass production in a microfluidic channel. The proposed microrobot is fabricated in a flow-focusing droplet generator by shearing a mixture of GelMA, photoinitiator, and superparamagnetic iron oxide nanoparticles (SPIONs) with a mixture of oil and surfactant. Human nasal turbinate stem cells (hNTSCs) are loaded on the GelMA microrobot, and the hNTSC-loaded microrobot shows precise rolling motion in response to an external rotating magnetic field. The microrobot is enzymatically degraded by collagenase, and released hNTSCs are proliferated and differentiated into neuronal cells. In addition, the feasibility of the GelMA microrobot as a cell therapeutic delivery system is investigated by measuring electrophysiological activity on a multielectrode array. Such a versatile and fully biodegradable microrobot has the potential for targeted stem cell delivery, proliferation, and differentiation for stem cell-based therapy. |
  | Sato Yuya; Yamamoto, Hideaki; Kato Hideyuki; Tanii Takashi; Sato Shigeo; Hirano-Iwata Ayumi Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks Journal Article 2022. @article{Sato2022, title = {Microfluidic cell engineering on high-density microelectrode arrays for assessing structure-function relationships in living neuronal networks}, author = {Sato, Yuya; Yamamoto, Hideaki; Kato, Hideyuki; Tanii, Takashi; Sato, Shigeo; Hirano-Iwata, Ayumi}, url = {https://arxiv.org/abs/2205.04342}, year = {2022}, date = {2022-05-11}, abstract = {Neuronal networks in dissociated culture combined with cell engineering technology offer a pivotal platform to constructively explore the relationship between structure and function in living neuronal networks. Here, we fabricated defined neuronal networks possessing a modular architecture on high-density microelectrode arrays (HD-MEAs), a state-of-the-art electrophysiological tool for recording neural activity with high spatial and temporal resolutions. We first established a surface coating protocol using a cell-permissive hydrogel to stably attach polydimethylsiloxane microfluidic film on the HD-MEA. We then recorded the spontaneous neural activity of the engineered neuronal network, which revealed an important portrait of the engineered neuronal network--modular architecture enhances functional complexity by reducing the excessive neural correlation between spatially segregated modules. The results of this study highlight the impact of HD-MEA recordings combined with cell engineering technologies as a novel tool in neuroscience to constructively assess the structure-function relationships in neuronal networks.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Neuronal networks in dissociated culture combined with cell engineering technology offer a pivotal platform to constructively explore the relationship between structure and function in living neuronal networks. Here, we fabricated defined neuronal networks possessing a modular architecture on high-density microelectrode arrays (HD-MEAs), a state-of-the-art electrophysiological tool for recording neural activity with high spatial and temporal resolutions. We first established a surface coating protocol using a cell-permissive hydrogel to stably attach polydimethylsiloxane microfluidic film on the HD-MEA. We then recorded the spontaneous neural activity of the engineered neuronal network, which revealed an important portrait of the engineered neuronal network--modular architecture enhances functional complexity by reducing the excessive neural correlation between spatially segregated modules. The results of this study highlight the impact of HD-MEA recordings combined with cell engineering technologies as a novel tool in neuroscience to constructively assess the structure-function relationships in neuronal networks. |
  | Shimba Kenta; Asahina, Takahiro; Sakai Koji; Kotani Kiyoshi; Jimbo Yasuhiko Recording Saltatory Conduction Along Sensory Axons Using a High-Density Microelectrode Array Journal Article Frontiers in Neuroscience, 2022. @article{Shimba2022, title = {Recording Saltatory Conduction Along Sensory Axons Using a High-Density Microelectrode Array}, author = {Shimba, Kenta; Asahina, Takahiro; Sakai, Koji; Kotani, Kiyoshi; Jimbo, Yasuhiko}, url = {https://www.frontiersin.org/articles/10.3389/fnins.2022.854637/full}, year = {2022}, date = {2022-04-18}, journal = {Frontiers in Neuroscience}, abstract = {In bottom-up neuroscience, questions on neural information processing are addressed by engineering small but reproducible biological neural networks of defined network topology in vitro. The network topology can be controlled by culturing neurons within polydimethylsiloxane (PDMS) microstructures that are combined with microelectrode arrays (MEAs) for electric access to the network. However, currently used glass MEAs are limited to 256 electrodes and pose a limitation to the spatial resolution as well as the design of more complex microstructures. The use of high density complementary metal-oxide-semiconductor (CMOS) MEAs greatly increases the spatial resolution, enabling sub-cellular readout and stimulation of neurons in defined neural networks. Unfortunately, the non-planar surface of CMOS MEAs complicates the attachment of PDMS microstructures. To overcome the problem of axons escaping the microstructures through the ridges of the CMOS MEA, we stamp-transferred a thin film of hexane-diluted PDMS onto the array such that the PDMS filled the ridges at the contact surface of the microstructures without clogging the axon guidance channels. This method resulted in 23 % of structurally fully connected but sealed networks on the CMOS MEA of which about 45 % showed spiking activity in all channels. Moreover, we provide an impedance-based method to visualize the exact location of the microstructures on the MEA and show that our method can confine axonal growth within the PDMS microstructures. Finally, the high spatial resolution of the CMOS MEA enabled us to show that action potentials follow the unidirectional topology of our circular multi-node microstructure.}, keywords = {}, pubstate = {published}, tppubtype = {article} } In bottom-up neuroscience, questions on neural information processing are addressed by engineering small but reproducible biological neural networks of defined network topology in vitro. The network topology can be controlled by culturing neurons within polydimethylsiloxane (PDMS) microstructures that are combined with microelectrode arrays (MEAs) for electric access to the network. However, currently used glass MEAs are limited to 256 electrodes and pose a limitation to the spatial resolution as well as the design of more complex microstructures. The use of high density complementary metal-oxide-semiconductor (CMOS) MEAs greatly increases the spatial resolution, enabling sub-cellular readout and stimulation of neurons in defined neural networks. Unfortunately, the non-planar surface of CMOS MEAs complicates the attachment of PDMS microstructures. To overcome the problem of axons escaping the microstructures through the ridges of the CMOS MEA, we stamp-transferred a thin film of hexane-diluted PDMS onto the array such that the PDMS filled the ridges at the contact surface of the microstructures without clogging the axon guidance channels. This method resulted in 23 % of structurally fully connected but sealed networks on the CMOS MEA of which about 45 % showed spiking activity in all channels. Moreover, we provide an impedance-based method to visualize the exact location of the microstructures on the MEA and show that our method can confine axonal growth within the PDMS microstructures. Finally, the high spatial resolution of the CMOS MEA enabled us to show that action potentials follow the unidirectional topology of our circular multi-node microstructure. |
  | Hornauer Philipp; Prack, Gustavo; Anastasi Nadia; Ronchi Silvia; Kim Taehoon; Donner Christian; Fiscella Michele; Borgwardt Karsten; Taylor Verdon; Jagasia Ravi; Roqueiro Damian; Hierlemann Andreas; Downregulating α-synuclein in iPSC-derived dopaminergic neurons mimics 2 electrophysiological phenotype of the A53T mutation Journal Article bioRxiv, 2022. @article{Hornauer2022, title = {Downregulating α-synuclein in iPSC-derived dopaminergic neurons mimics 2 electrophysiological phenotype of the A53T mutation}, author = {Hornauer, Philipp; Prack, Gustavo; Anastasi, Nadia; Ronchi, Silvia; Kim, Taehoon; Donner, Christian; Fiscella, Michele; Borgwardt, Karsten; Taylor, Verdon; Jagasia, Ravi; Roqueiro, Damian; Hierlemann, Andreas;}, url = {https://www.biorxiv.org/content/10.1101/2022.03.31.486582v1.full}, year = {2022}, date = {2022-04-01}, journal = {bioRxiv}, abstract = {Parkinson’s disease (PD) is a common debilitating neurodegenerative disorder, characterized by a progressive loss of dopaminergic (DA) neurons. Mutations, gene dosage increase, and single nucleotide polymorphisms in the α-synuclein-encoding gene SNCA either cause or increase the risk for PD. However, neither the function of α-synuclein in health and disease, nor its role throughout development is fully understood. Here, we introduce DeePhys, a new tool that allows for data-driven functional phenotyping of neuronal cell lines by combining electrophysiological features inferred from high-density microelectrode array (HD-MEA) recordings with a robust machine learning workflow. We apply DeePhys to human induced pluripotent stem cell (iPSC)-derived DA neuron-astrocyte co-cultures harboring the prominent SNCA mutation A53T and an isogenic control line. Moreover, we demonstrate how DeePhys can facilitate the assessment of cellular and network-level electrophysiological features to build functional phenotypes and to evaluate potential treatment interventions. We find that electrophysiological features across all scales proved to be highly specific for the A53T phenotype, enabled to predict the genotype and age of individual cultures with high accuracy, and revealed a mutant-like phenotype after downregulation of α-synuclein.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Parkinson’s disease (PD) is a common debilitating neurodegenerative disorder, characterized by a progressive loss of dopaminergic (DA) neurons. Mutations, gene dosage increase, and single nucleotide polymorphisms in the α-synuclein-encoding gene SNCA either cause or increase the risk for PD. However, neither the function of α-synuclein in health and disease, nor its role throughout development is fully understood. Here, we introduce DeePhys, a new tool that allows for data-driven functional phenotyping of neuronal cell lines by combining electrophysiological features inferred from high-density microelectrode array (HD-MEA) recordings with a robust machine learning workflow. We apply DeePhys to human induced pluripotent stem cell (iPSC)-derived DA neuron-astrocyte co-cultures harboring the prominent SNCA mutation A53T and an isogenic control line. Moreover, we demonstrate how DeePhys can facilitate the assessment of cellular and network-level electrophysiological features to build functional phenotypes and to evaluate potential treatment interventions. We find that electrophysiological features across all scales proved to be highly specific for the A53T phenotype, enabled to predict the genotype and age of individual cultures with high accuracy, and revealed a mutant-like phenotype after downregulation of α-synuclein. |
  | Sawada, Haruto; Wake, Naoki; Sasabuchi, Kazuhiro; Takamatsu, Jun; Takahashi, Hirokazu; Ikeuchi, Katsushi Design strategies for controlling neuron-connected robots using reinforcement learning Journal Article arXiv, 2022. @article{Sawada2022, title = {Design strategies for controlling neuron-connected robots using reinforcement learning}, author = {Haruto Sawada and Naoki Wake and Kazuhiro Sasabuchi and Jun Takamatsu and Hirokazu Takahashi and Katsushi Ikeuchi}, url = {https://arxiv.org/abs/2203.15290}, doi = {https://doi.org/10.48550/arXiv.2203.15290}, year = {2022}, date = {2022-03-29}, journal = {arXiv}, abstract = {Despite the growing interest in robot control utilizing the computation of biological neurons, context-dependent behavior by neuron-connected robots remains a challenge. Context-dependent behavior here is defined as behavior that is not the result of a simple sensory-motor coupling, but rather based on an understanding of the task goal. This paper proposes design principles for training neuron-connected robots based on task goals to achieve context-dependent behavior. First, we employ deep reinforcement learning (RL) to enable training that accounts for goal achievements. Second, we propose a neuron simulator as a probability distribution based on recorded neural data, aiming to represent physiologically valid neural dynamics while avoiding complex modeling with high computational costs. Furthermore, we propose to update the simulators during the training to bridge the gap between the simulation and the real settings. The experiments showed that the robot gradually learned context-dependent behaviors in pole balancing and robot navigation tasks. Moreover, the learned policies were valid for neural simulators based on novel neural data, and the task performance increased by updating the simulators during training. These results suggest the effectiveness of the proposed design principle for the context-dependent behavior of neuron-connected robots.}, keywords = {}, pubstate = {published}, tppubtype = {article} } Despite the growing interest in robot control utilizing the computation of biological neurons, context-dependent behavior by neuron-connected robots remains a challenge. Context-dependent behavior here is defined as behavior that is not the result of a simple sensory-motor coupling, but rather based on an understanding of the task goal. This paper proposes design principles for training neuron-connected robots based on task goals to achieve context-dependent behavior. First, we employ deep reinforcement learning (RL) to enable training that accounts for goal achievements. Second, we propose a neuron simulator as a probability distribution based on recorded neural data, aiming to represent physiologically valid neural dynamics while avoiding complex modeling with high computational costs. Furthermore, we propose to update the simulators during the training to bridge the gap between the simulation and the real settings. The experiments showed that the robot gradually learned context-dependent behaviors in pole balancing and robot navigation tasks. Moreover, the learned policies were valid for neural simulators based on novel neural data, and the task performance increased by updating the simulators during training. These results suggest the effectiveness of the proposed design principle for the context-dependent behavior of neuron-connected robots. |
  | Duru Jens; Küchler, Joël; Ihle Stephan Forró Csaba; Bernardi Aeneas; Girardin Sophie; Hengsteler Julian; Wheeler Stephen; János; Ruff Tobias; J ; Vörös Engineered Biological Neural Networks on High Density CMOS Microelectrode Arrays Journal Article 2022. @article{Duru2022, title = {Engineered Biological Neural Networks on High Density CMOS Microelectrode Arrays}, author = {Duru, Jens; Küchler, Joël; Ihle, Stephan J.;, Forró, Csaba; Bernardi, Aeneas; Girardin, Sophie; Hengsteler, Julian; Wheeler, Stephen; Vörös, János; Ruff, Tobias;}, url = {https://www.frontiersin.org/articles/10.3389/fnins.2022.829884/full}, year = {2022}, date = {2022-02-21}, abstract = {In bottom-up neuroscience, questions on neural information processing are addressed by engineering small but reproducible biological neural networks of defined network topology in vitro. The network topology can be controlled by culturing neurons within polydimethylsiloxane (PDMS) microstructures that are combined with microelectrode arrays (MEAs) for electric access to the network. However, currently used glass MEAs are limited to 256 electrodes and pose a limitation to the spatial resolution as well as the design of more complex microstructures. The use of high density complementary metal-oxide-semiconductor (CMOS) MEAs greatly increases the spatial resolution, enabling sub-cellular readout and stimulation of neurons in defined neural networks. Unfortunately, the non-planar surface of CMOS MEAs complicates the attachment of PDMS microstructures. To overcome the problem of axons escaping the microstructures through the ridges of the CMOS MEA, we stamp-transferred a thin film of hexane-diluted PDMS onto the array such that the PDMS filled the ridges at the contact surface of the microstructures without clogging the axon guidance channels. This method resulted in 23 % of structurally fully connected but sealed networks on the CMOS MEA of which about 45 % showed spiking activity in all channels. Moreover, we provide an impedance-based method to visualize the exact location of the microstructures on the MEA and show that our method can confine axonal growth within the PDMS microstructures. Finally, the high spatial resolution of the CMOS MEA enabled us to show that action potentials follow the unidirectional topology of our circular multi-node microstructure.}, keywords = {}, pubstate = {published}, tppubtype = {article} } In bottom-up neuroscience, questions on neural information processing are addressed by engineering small but reproducible biological neural networks of defined network topology in vitro. The network topology can be controlled by culturing neurons within polydimethylsiloxane (PDMS) microstructures that are combined with microelectrode arrays (MEAs) for electric access to the network. However, currently used glass MEAs are limited to 256 electrodes and pose a limitation to the spatial resolution as well as the design of more complex microstructures. The use of high density complementary metal-oxide-semiconductor (CMOS) MEAs greatly increases the spatial resolution, enabling sub-cellular readout and stimulation of neurons in defined neural networks. Unfortunately, the non-planar surface of CMOS MEAs complicates the attachment of PDMS microstructures. To overcome the problem of axons escaping the microstructures through the ridges of the CMOS MEA, we stamp-transferred a thin film of hexane-diluted PDMS onto the array such that the PDMS filled the ridges at the contact surface of the microstructures without clogging the axon guidance channels. This method resulted in 23 % of structurally fully connected but sealed networks on the CMOS MEA of which about 45 % showed spiking activity in all channels. Moreover, we provide an impedance-based method to visualize the exact location of the microstructures on the MEA and show that our method can confine axonal growth within the PDMS microstructures. Finally, the high spatial resolution of the CMOS MEA enabled us to show that action potentials follow the unidirectional topology of our circular multi-node microstructure. |
Discover More
Would you like to learn more about MaxOne? Book a one-to-one call with one of our scientist to discuss how MaxWell Biosystems’ high-content electrophysiology solutions can bring new key insights to your project or request a quote.
Support
We provide training and support on all aspects of the MaxTwo platform. We have the expertise to help you design experiments and/or analysis tools and integrate MaxTwo with your automation system.
Ask a Question English
English


